Bridging the Gap: A Strategic Guide to In Vitro and In Vivo Biomaterial Testing Comparison
This article provides a comprehensive analysis for researchers and drug development professionals on the critical comparison between in vitro and in vivo biomaterial testing.
Bridging the Gap: A Strategic Guide to In Vitro and In Vivo Biomaterial Testing Comparison
Abstract
This article provides a comprehensive analysis for researchers and drug development professionals on the critical comparison between in vitro and in vivo biomaterial testing. It explores the foundational principles of both methodologies, detailing their specific applications as guided by standards like ISO 10993. The content addresses common challenges, such as the poor correlation between test systems, and presents optimization strategies, including advanced 3D models and refined test conditions, to enhance predictive accuracy. Finally, it establishes a framework for the synergistic use of both testing paradigms to validate biocompatibility and safety, ensuring a more efficient and ethically conscious path from laboratory research to clinical application.
Understanding the Basics: Defining In Vitro and In Vivo Testing Paradigms
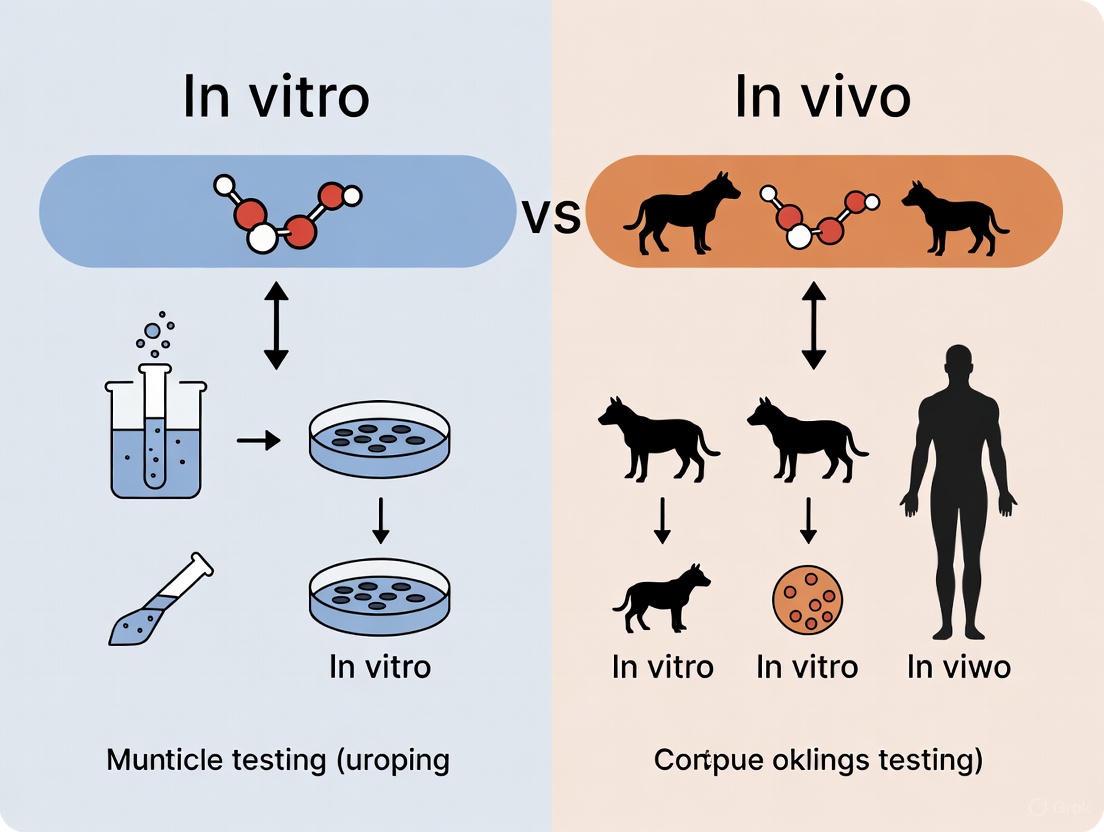
In biomaterial science, predicting how a material will behave once implanted in the human body is the central challenge. Research in this field rests on two fundamental methodological pillars: in vitro and in vivo studies. The term in vitro, Latin for "in glass," refers to experiments performed with microorganisms, cells, or biological molecules outside their normal biological context, typically in labware like Petri dishes or test tubes [1] [2]. In contrast, in vivo, Latin for "within the living," describes studies conducted in or on a whole living organism, such as a laboratory animal or human [3] [2]. The core objective for scientists is to translate findings from the controlled simplicity of in vitro assays to the complex, dynamic reality of an in vivo environment. This guide provides a detailed comparison of these approaches, focusing on their application in evaluating biomaterials for soft tissue reconstruction, to help researchers navigate the strengths and limitations of each.
Core Conceptual Definitions and Comparisons
Defining the Paradigms
In Vitro studies are characterized by their use of components isolated from a living organism. This methodology permits a more detailed or convenient analysis than can be done with whole organisms and is colloquially known as "test-tube experiments" [1]. These studies simplify the system under investigation, allowing researchers to focus on a small number of components, such as the interaction between a specific cell type and a biomaterial [1]. A key advantage is species specificity; human cells can be studied directly, eliminating the need to extrapolate from animal models [1]. Furthermore, in vitro methods are relatively inexpensive, fast, and can be miniaturized and automated for high-throughput screening [1] [4].
In Vivo studies, on the other hand, are defined by their use of a whole, living organism. These studies are designed to provide information regarding the effects of a substance or disease progression in a complex, physiologically complete system [3] [2]. In vivo work is essential for assessing the safety, efficacy, and overall therapeutic profile of a drug or biomaterial before human use [3] [4]. It captures the intricate interplay between different organs, tissues, and physiological factors that cannot be replicated in a dish [5]. However, these studies are very expensive, time-consuming, and are subject to strict ethical regulations [4].
Comparative Analysis at a Glance
Table 1: A side-by-side comparison of in vivo and in vitro methodologies.
| Aspect | In Vitro | In Vivo |
|---|---|---|
| Definition | Studies performed "in the glass" outside a living organism [1] [4]. | Studies performed "within a living" organism [3] [4]. |
| System Complexity | Simplified, controlled environment using isolated cells or tissues [1] [5]. | Complex, fully integrated physiological system [5]. |
| Cost & Duration | Relatively low cost and fast results [4]. | Very expensive and long-term studies [4]. |
| Primary Applications | Initial drug/biomaterial screening, mechanism of action studies, high-throughput assays [1] [4]. | Preclinical safety and efficacy testing, pharmacokinetics/pharmacodynamics (PK/PD), clinical trials [3] [4]. |
| Key Advantages | Experimental control, species specificity, convenience, automation [1] [4]. | Provides specific and reliable data on biological effects in a whole organism [4]. |
| Key Limitations | Physiologically limited; results may not predict in vivo outcomes [1] [4]. | Strict ethical regulations; high cost and complexity; interspecies differences [4] [5]. |
Experimental Models and Methodologies
In Vitro Model Systems
Traditional in vitro models often involve two-dimensional (2D) cell cultures, where a single cell type is grown on a flat plastic surface. While useful for foundational studies, 2D cultures are limited as they cannot replicate the three-dimensional architecture or cell-to-cell interactions found in living tissues [6]. To bridge this gap, Complex In Vitro Models (CIVMs) have been developed.
CIVMs are systems that integrate a multicellular environment within a three-dimensional (3D) biopolymer or tissue-derived matrix [6]. These models seek to reconstruct organ- or tissue-specific characteristics to better mimic in vivo conditions [6]. Key CIVM technologies include:
- Organoids: 3D structures derived from stem cells that self-organize into organ-like structures, recapitulating some functional aspects of the source organ [6].
- Organs-on-a-Chip: Microfluidic devices that combine cell culture with biomedical engineering to simulate a physiological organ environment, including mechanical forces like fluid flow and stretch [6] [4].
- Microphysiological Systems (MPS): Interconnected constructs that combine tissue engineering and biosensors to emulate the functions of an organ in vitro [4].
In Vivo Model Systems
In vivo testing in biomaterial and drug development typically proceeds in two main stages:
- Animal Models (Preclinical): Before human testing, a biomaterial must undergo rigorous evaluation in animals to assess its toxicity, biocompatibility, and efficacy [3] [4]. Rodents, particularly mice, are the most commonly used species due to their size, cost, fast reproduction, and genetic similarity to humans [4]. Studies are designed using disease models induced through invasive procedures, administration of drugs/toxicants, or genetic alteration [3].
- Human Trials (Clinical): If a biomaterial or drug proves safe and effective in animal studies, it progresses to clinical trials in humans [2] [4]. These trials are tightly controlled and monitored to evaluate the safety and efficacy of the intervention in human subjects, often using randomized and blinded designs to ensure result integrity [2].
Quantitative Data: Comparing In Vitro and In Vivo Outcomes
A critical challenge in biomaterial science is correlating in vitro findings with in vivo outcomes. The following data, derived from a study on the host response to implanted materials, illustrates this point.
Table 2: Quantitative comparison of in vitro macrophage responses and in vivo remodeling outcomes for various biomaterials. Data adapted from a study linking in vitro assays to in vivo outcomes [7].
| Material | In Vitro Macrophage Secretion (Relative Levels) | Predicted In Vivo Response (from in vitro data) | Actual In Vivo Remodeling Score (14 & 35 days) | Dominant In Vivo Macrophage Phenotype |
|---|---|---|---|---|
| MatriStem (ECM) | Distinct anti-inflammatory profile | Constructive Remodeling | High | M2 (tissue remodeling) |
| CL-MatriStem (Crosslinked ECM) | Pro-inflammatory profile | Foreign Body Reaction | Low | M1 (pro-inflammatory) |
| D-ECM (Dermal ECM) | Distinct anti-inflammatory profile | Constructive Remodeling | High | M2 (tissue remodeling) |
| Vicryl (Synthetic) | Pro-inflammatory profile | Foreign Body Reaction | Low | M1 (pro-inflammatory) |
| Dual Mesh (Synthetic) | Pro-inflammatory profile | Foreign Body Reaction | Low | M1 (pro-inflammatory) |
Key Findings from the Data:
- ECM-based materials (MatriStem, D-ECM), which are naturally derived, consistently prompted a favorable anti-inflammatory response in vitro that accurately predicted constructive remodeling and a dominant M2 macrophage presence in vivo [7].
- Synthetic and crosslinked materials (Vicryl, Dual Mesh, CL-MatriStem) elicited a pro-inflammatory response in vitro, which correlated with a foreign body reaction, lower remodeling scores, and a dominant M1 macrophage phenotype in vivo [7].
- This study demonstrated that when combined with sophisticated in silico analysis (e.g., Principal Component Analysis and Dynamic Network Analysis), in vitro macrophage assays can serve as a valuable predictor of the in vivo host response to implanted biomaterials [7].
Detailed Experimental Protocols
A Representative In Vitro Protocol: Macrophage-Biomaterial Assay
This protocol is designed to evaluate the inflammatory potential of a biomaterial by characterizing the dynamic response of human macrophages [7].
Objective: To assess the activation profile of human monocyte-derived macrophages exposed to various biomaterials and use the cytokine secretion profile to predict the in vivo remodeling outcome.
Materials and Reagents:
- Test Materials: Synthetic and biologically derived materials (e.g., ECM scaffolds, polymer meshes) cut into standardized discs.
- Cells: Human monocytes isolated from peripheral blood and differentiated into macrophages.
- Cell Culture Medium: RPMI-1640 supplemented with fetal bovine serum, antibiotics, and macrophage colony-stimulating factor (M-CSF) for differentiation.
- Analysis Kits: ELISA kits for quantifying cytokine secretion (e.g., IL-1β, IL-6, IL-8, IL-10, TNF-α).
Methodology:
- Material Sterilization: All material discs are terminally sterilized using ethylene oxide gas or other appropriate methods [7].
- Macrophage Differentiation: Isolated human monocytes are cultured in M-CSF-containing medium for 7 days to differentiate them into macrophages.
- Material Exposure: Differentiated macrophages are seeded onto the test material discs placed in multi-well culture plates. Control wells contain cells without material or with a reference material.
- Conditioned Media Collection: The culture medium (conditioned media) is collected at multiple time points (e.g., 24, 48, 72 hours) to capture dynamic cytokine secretion.
- Cytokine Quantification: Collected media is analyzed using ELISA to determine the concentrations of key pro-inflammatory (e.g., IL-1β, TNF-α) and anti-inflammatory (e.g., IL-10) cytokines.
- In Silico Analysis: The multidimensional cytokine data is processed using computational methods like Principal Component Analysis (PCA) to identify distinct response patterns and predict the in vivo outcome [7].
A Representative In Vivo Protocol: Rodent Abdominal Wall Defect Model
This established model is used to evaluate the host remodeling response to a biomaterial in a clinically relevant soft tissue location [7].
Objective: To implant material coupons into a rodent model and assess the in vivo host response, including tissue remodeling and immune cell infiltration.
Materials and Reagents:
- Animals: Female Sprague-Dawley rats.
- Test Materials: Material coupons (e.g., 1x1 cm).
- Surgical Supplies: Sutures (non-degradable for fixation, degradable for skin closure), anesthetic agents, standard surgical instruments.
- Histology Reagents: Neutral buffered formalin, paraffin, hematoxylin and eosin (H&E) stain, antibodies for immunofluorescence (e.g., for M1/M2 macrophage markers).
Methodology:
- Surgical Implantation: A paramedian partial-thickness abdominal wall defect is created in an anesthetized rat. The external and internal oblique muscles are excised, leaving the transversalis fascia intact [7].
- Graft Placement: The size-matched material coupon is inlaid within the defect and fixed to the adjacent muscle at the four corners with non-degradable sutures.
- Post-Op and Explant: The skin incision is closed, and animals are monitored post-operatively. After the predetermined implantation period (e.g., 14 and 35 days), the animals are euthanized, and the implants with surrounding tissue are explanted.
- Histological Processing: Explanted tissues are fixed in formalin, dehydrated, embedded in paraffin, and sectioned into thin slices for staining [7].
- Histological Analysis:
- H&E Staining: Used for general histomorphology. A blinded observer scores the tissues semiquantitatively for foreign body giant cell formation, connective tissue organization, encapsulation, and muscle ingrowth to generate a total remodeling score [7].
- Immunofluorescent Staining: Used to identify macrophage phenotype. Sections are labeled with antibodies against M1 markers (e.g., CCR7) and M2 markers (e.g., CD206) to determine the dominant immune response [7].
Visualization of Workflows and Relationships
The In Vitro to In Vivo Translation Workflow
The following diagram illustrates the sequential and iterative process of translating findings from basic in vitro models to predictive complex models and finally to in vivo validation.
Diagram 1: The iterative workflow for translating in vitro findings to in vivo outcomes.
The Host Response to Biomaterials
This diagram contrasts the typical biological pathways activated by a pro-inflammatory biomaterial versus a constructive remodeling biomaterial, linking cellular events to tissue-level outcomes.
Diagram 2: Key signaling pathways in the host response to biomaterials.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key reagents, materials, and technologies used in biomaterial testing.
| Tool | Category | Primary Function in Research |
|---|---|---|
| Petri Dishes / Multi-well Plates | Labware | Provide a sterile, controlled environment for 2D cell culture and in vitro assays [1]. |
| Extracellular Matrix (ECM) Scaffolds | Biomaterial | Naturally derived materials (e.g., MatriStem) used as test substrates or as components of CIVMs to provide a physiological 3D environment [6] [7]. |
| Synthetic Polymer Meshes | Biomaterial | Materials (e.g., Polypropylene, ePTFE, Vicryl) used as test substrates to study the foreign body reaction and synthetic graft integration [7]. |
| ELISA Kits | Analytical Tool | Quantify specific protein secretion (e.g., cytokines) in in vitro conditioned media or in vivo tissue/tuid samples to assess immune response [7]. |
| Primary Cells (e.g., Macrophages) | Biological Tool | Isolated directly from tissue (human or animal); used in in vitro assays for more physiologically relevant responses than cell lines [7]. |
| Organ-on-a-Chip Devices | CIVM Technology | Microfluidic devices that simulate organ-level physiology for more predictive in vitro drug and toxicology testing [6] [4]. |
| Immunofluorescence Antibodies | Analytical Tool | Allow visualization and identification of specific cell types (e.g., M1/M2 macrophages) and structures in tissue sections from in vivo studies [7]. |
The journey from in vitro discovery to in vivo application remains a central challenge in biomaterial science. While in vitro models offer unparalleled control and throughput for initial screening, their physiological limitations mean results cannot be directly extrapolated to predict outcomes in a living organism [1] [8]. In vivo studies are indispensable for validating safety and efficacy in a whole-body context, but their cost and complexity necessitate their judicious use [4] [5]. The future of the field lies in the development and adoption of more sophisticated Complex In Vitro Models (CIVMs), such as organs-on-chips and patient-derived organoids, which better mimic human physiology [6]. When combined with powerful in silico modeling, these advanced in vitro systems are poised to improve predictive accuracy, help down-select the most promising candidates for in vivo studies, and ultimately accelerate the development of safer and more effective biomaterials [7] [5].
The Critical Role of Testing in the Biomaterial Development Pipeline
The development of novel biomaterials is a complex, multi-stage process where rigorous testing serves as the critical gateway between laboratory innovation and clinical application. Biomaterials, defined as materials engineered to interact with biological systems for medical or therapeutic purposes, must demonstrate exceptional safety, efficacy, and reliability before human use [9]. The testing pathway strategically employs complementary methodologies—in vitro (in glass) and in vivo (within the living) models—to comprehensively evaluate material performance [2] [10]. In vitro studies occur in controlled laboratory environments, such as test tubes or petri dishes, and are ideal for initial screening and mechanistic studies. In contrast, in vivo studies are conducted within whole living organisms, providing essential data on complex physiological interactions and systemic responses [11]. This article objectively compares the performance data, applications, and limitations of these testing paradigms, providing researchers and drug development professionals with a clear framework for navigating the biomaterial development pipeline.
Fundamental Principles: In Vitro vs. In Vivo Testing
Understanding the core definitions and philosophical approaches of each testing methodology is foundational to designing an effective development strategy.
In Vitro Testing: Latin for "in glass," this approach involves experiments performed outside of a living organism [2]. Cells or tissues are maintained in highly controlled, artificial environments, such as culture plates or bioreactors [10]. The primary advantage of this method is the high degree of control over experimental variables, which allows for precise, cost-effective, and rapid screening of biomaterial properties, including cytotoxicity and initial biocompatibility [11] [12]. However, its significant limitation is the inability to replicate the full complexity of a living system, including immune responses, organ-level interactions, and metabolic processes [10] [12].
In Vivo Testing: Latin for "within the living," this methodology involves testing within a whole living organism, typically animal models such as rodents or non-human primates [2] [10]. This approach provides the most physiologically relevant data on how a biomaterial interacts with a complete biological system, informing aspects like long-term biocompatibility, integration with host tissues, and overall safety profile [11]. The main drawbacks include high costs, lengthy timelines, ethical considerations regarding animal use, and greater variability due to inter-individual differences [10] [11].
Table 1: Core Conceptual Differences Between In Vitro and In Vivo Testing.
| Aspect | In Vitro Testing | In Vivo Testing |
|---|---|---|
| Definition | Experiments conducted outside a living organism [2] | Experiments conducted within a living organism [2] |
| Experimental Model | Cell cultures, tissues, or organs in an artificial environment [12] | Whole, living animals (e.g., rodents, primates) [10] |
| Key Advantage | High control over variables; cost-effective and rapid [11] | Provides full physiological and systemic context [11] |
| Primary Limitation | Lacks the complexity of a full organism [10] | High cost, time-consuming, and raises ethical concerns [11] |
Performance Data and Comparative Analysis
The choice between in vitro and in vivo models directly influences the type and reliability of performance data obtained for a biomaterial. The key performance parameters in pre-clinical assessment include mechanical integrity, biocompatibility, and degradation behavior [13].
Quantitative Performance Comparison
Data from the search results allows for a direct comparison of how these two testing methods are used to evaluate critical biomaterial properties.
Table 2: Comparative Analysis of Biomaterial Performance Assessment In Vitro vs. In Vivo.
| Performance Parameter | In Vitro Testing Methods & Findings | In Vivo Testing Methods & Findings |
|---|---|---|
| Biocompatibility & Host Response | Macrophage Activation Assays: Cytokine secretion profiles predict foreign body reaction vs. constructive remodeling [7]. | Rodent Implantation Models: Histology shows crosslinked ECM/synthetics cause fibrosis; non-crosslinked ECM promotes constructive remodeling [7]. |
| Mechanical Properties | Tensile Testing: PLA and PHA show sufficient strength and flexibility for load-bearing tissue engineering [9]. | Not the primary method for assessment. Mechanical performance is inferred from implant integrity and functional recovery in the host. |
| Degradation Behavior | Chemical Analysis & Accelerated Aging: Studies on PLA, PHA, and chitosan elucidate degradation rates in controlled biochemical environments [9]. | Explants & Histology: Reveals real-time degradation rate, clearance of byproducts, and correlation between degradation and tissue healing [7]. |
Correlation and Predictive Value
A central challenge in biomaterial development is the translational gap between simplified in vitro models and complex in vivo outcomes. While in vitro data is precise, it does not always accurately predict in vivo responses [12]. For instance, a significant number of drug candidates fail in human clinical trials despite promising pre-clinical in vitro and animal data [2]. To bridge this gap, advanced in vitro models are evolving. For example, one study developed a sophisticated in vitro assay using human macrophages, which, when combined with in silico analysis, successfully predicted the in vivo host response (constructive remodeling versus foreign body reaction) to various biomaterials in a rodent model [7]. Furthermore, technologies like Organ-on-a-Chip systems, which create more physiologically relevant microenvironments with human cells, are emerging as powerful tools to improve predictive accuracy and reduce reliance on animal models [12].
Experimental Protocols and Methodologies
Detailed and standardized protocols are essential for generating reproducible and meaningful data in biomaterial testing. Below are detailed methodologies for key experiments cited in this field.
Detailed Protocol: In Vitro Macrophage Activation Assay
This protocol, adapted from a foundational study, is used to predict the in vivo host response to biomaterials by characterizing the dynamic inflammatory response of macrophages [7].
- Objective: To characterize the dynamic inflammatory response of human monocyte-derived-macrophages to biomaterials and identify secretory profiles predictive of in vivo remodeling outcomes.
- Materials Preparation:
- Biomaterial Samples: Cut synthetic and ECM-derived materials (e.g., Polypropylene, Decellularized ECM) into standardized discs (e.g., 2-cm diameter).
- Sterilization: Terminally sterilize all devices using ethylene oxide gas or other appropriate methods that do not alter material properties.
- Cell Culture: Isulate human primary monocytes from peripheral blood and differentiate them into macrophages using Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF) over 7 days.
- Methodology:
- Culture Setup: Place sterilized biomaterial discs in multi-well culture plates. Seed differentiated macrophages onto the material surfaces at a standardized density (e.g., 500,000 cells/well) in serum-free medium.
- Conditioned Media Collection: Collect cell-conditioned culture media at multiple time points (e.g., 1, 3, 5, and 7 days).
- Analysis of Secreted Factors: Analyze the conditioned media using multiplex immunoassays (e.g., Luminex) to quantify the concentration of key pro-inflammatory (e.g., TNF-α, IL-1β, IL-6) and pro-remodeling (e.g., IL-10, TGF-β) cytokines and chemokines.
- In Silico Analysis: Subject the multi-analyte, time-course data to quasi-mechanistic in silico analysis, such as Principal Component Analysis (PCA) and Dynamic Network Analysis (DyNA), to identify dominant, predictive patterns in the macrophage response.
- Outputs: PCA and DyNA generate distinct activation profiles that can discriminate between materials that will elicit a pro-inflammatory (M1) response versus a pro-remodeling (M2) response in vivo [7].
Detailed Protocol: In Vivo Rodent Biocompatibility and Remodeling
This protocol assesses the functional host response and tissue integration of a biomaterial in a live, complex physiological environment [7].
- Objective: To evaluate the in vivo host remodeling response and macrophage polarization to an implanted biomaterial in a rodent skeletal muscle model.
- Materials Preparation:
- Implant Coupons: Cut test biomaterials into small, uniform coupons (e.g., 1 cm x 1 cm).
- Sterilization: Ensure all implants are terminally sterilized as for the in vitro assays.
- Animal Model: Utilize an established model, such as a partial-thickness abdominal wall defect in Sprague-Dawley rats.
- Surgical Implantation:
- Anesthesia and Asepsis: Anesthetize the rodent and prepare the surgical site using aseptic technique.
- Defect Creation: Create a standardized partial-thickness defect in the abdominal wall muscle.
- Implantation: Inlay the biomaterial coupon into the defect and fix it to the adjacent native tissue at the corners using non-degradable sutures for positional stability.
- Closure: Close the surgical site with degradable sutures.
- Post-Op and Analysis:
- Explanation: After a pre-determined implantation period (e.g., 14 and 35 days), euthanize the animals and explant the device with the surrounding host tissue.
- Histological Processing: Fix the explants in formalin, embed in paraffin, and section into thin slices (5 μm) for staining.
- Histological Scoring: Stain sections with Hematoxylin and Eosin (H&E) and score them semiquantitatively for key remodeling characteristics: foreign body giant cell formation, degradation, connective tissue organization, encapsulation, and muscle ingrowth. A higher total score indicates more favorable remodeling.
- Immunofluorescence Staining: Label tissue sections with antibodies for macrophage phenotype markers (e.g., CCR7 for M1 and CD206 for M2) to determine the dominant immune response at the implant site.
Research Reagent Solutions and Essential Materials
A successful testing pipeline relies on a suite of specialized reagents, equipment, and models. The following table details key solutions used in the featured experiments and the broader field.
Table 3: Essential Research Reagent Solutions for Biomaterial Testing.
| Item Name | Function/Application | Specific Example/Context |
|---|---|---|
| Universal Testing Machines [14] | Evaluate mechanical properties (tensile strength, compression, elasticity) of biomaterials. | Used for in vitro tensile testing of biopolymers like PLA and PHA to assess strength for load-bearing applications [9]. |
| Human Monocyte-Derived Macrophages [7] | Model the human innate immune response to biomaterials in vitro. | Central to the predictive macrophage activation assay; respond to materials by secreting characteristic cytokine profiles [7]. |
| Extracellular Matrix (ECM)-Derived Materials [7] | Biologically derived scaffolds (e.g., decellularized dermis, urinary bladder) used as benchmark materials or test articles. | MatriStem (urinary bladder ECM) is used in vivo and shown to promote constructive remodeling with an M2 macrophage presence [7]. |
| Dynamic Mechanical Analyzers & Rheometers [14] | Characterize viscoelastic properties and time-dependent mechanical behavior of materials. | Used to measure self-healing properties and strain-stiffening behavior of advanced hydrogels mimicking natural tissues [15]. |
| Rodent Skeletal Muscle Injury Model [7] | A standard in vivo model for assessing biocompatibility and functional tissue integration of implants. | Provides a robust platform for evaluating the host remodeling response to materials in a soft tissue location [7]. |
| Multiplex Immunoassays | Simultaneously quantify multiple soluble proteins (e.g., cytokines, chemokines) from cell culture media or tissue lysates. | Critical for analyzing the complex secretory profile of macrophages in the in vitro predictive assay [7]. |
Visualizing the Testing Pipeline and a Key Assay
To clarify the logical workflow of the biomaterial testing pipeline and the specifics of a key predictive assay, the following diagrams were generated using Graphviz.
The development of safe and effective biomaterials is unequivocally dependent on a critical, multi-staged testing pipeline that strategically integrates both in vitro and in vivo methodologies. As demonstrated by the comparative data and protocols, in vitro studies provide an unparalleled platform for high-throughput, cost-effective screening of fundamental material properties and initial biological interactions [11] [9]. Conversely, in vivo testing remains indispensable for validating these findings within the irreducible complexity of a whole living organism, revealing systemic responses and long-term functional outcomes that cannot be fully modeled in a dish [11] [7]. The evolving paradigm in the field is not to view these methods as competitors, but as powerfully complementary tools. The emergence of advanced in vitro systems like Organ-on-a-Chip and sophisticated in silico analytical techniques is actively bridging the translational gap, enhancing predictive power, and refining the path to clinical success [12] [7]. For researchers and drug development professionals, a nuanced understanding of the strengths, limitations, and appropriate applications of each testing approach is fundamental to efficiently navigating the biomaterial development pipeline and delivering innovative solutions to patients.
The ISO 10993 series, developed by the International Organization for Standardization (ISO), comprises a set of internationally recognized standards for the biological evaluation of medical devices [16]. These standards provide a systematic framework for assessing the potential biological risks posed by medical devices that come into direct or indirect contact with the human body [17] [18]. The fundamental principle underlying these standards is the evaluation of biocompatibility—defined as the "ability of a medical device or material to perform with an appropriate host response in a specific application" [19]. Compliance with ISO 10993 standards helps ensure that medical devices meet essential safety and performance criteria, thereby contributing to the overall quality and reliability of healthcare products while facilitating global market access [16] [18].
The evaluation process outlined in ISO 10993 is integrated within a risk management framework aligned with ISO 14971, the standard for application of risk management to medical devices [18] [20]. This integration emphasizes that biological evaluation should not be a mere checklist of tests but a science-based, risk-informed process conducted throughout the device lifecycle—from design and development to post-market surveillance [20]. The standards are utilized by medical device manufacturers when preparing various regulatory submissions, including Premarket Applications (PMAs), Investigational Device Exemption (IDE) Applications, and Premarket Notifications (510(k)s) in the United States, and are similarly recognized by other regulatory bodies worldwide [16] [17].
The ISO 10993 Series: A Framework for Biological Safety Assessment
The ISO 10993 series consists of multiple parts, each addressing specific aspects of biological safety evaluation. The following table provides an overview of the key standards within the series:
| Standard Number | Title and Focus Area | Latest Version |
|---|---|---|
| ISO 10993-1 | Evaluation and testing within a risk management process | 2025 (expected) [19] [20] |
| ISO 10993-2 | Animal welfare requirements | 2022 [16] |
| ISO 10993-3 | Tests for genotoxicity, carcinogenicity, and reproductive toxicity | 2014 [16] |
| ISO 10993-4 | Selection of tests for interactions with blood | 2017 [16] |
| ISO 10993-5 | Tests for in vitro cytotoxicity | 2009 [16] |
| ISO 10993-6 | Tests for local effects after implantation | 2016 [16] |
| ISO 10993-7 | Ethylene oxide sterilization residuals | 2008 [16] |
| ISO 10993-9 | Framework for identification and quantification of potential degradation products | 2019 [16] |
| ISO 10993-10 | Tests for skin sensitization | 2021 [16] |
| ISO 10993-11 | Tests for systemic toxicity | 2018 [16] |
| ISO 10993-12 | Sample preparation and reference materials | 2021 [16] [21] |
| ISO 10993-17 | Toxicological risk assessment of medical device constituents | 2023 [16] |
| ISO 10993-18 | Chemical characterization of medical device materials | 2023 [16] |
The Central Role of ISO 10993-1
ISO 10993-1 serves as the cornerstone document of the series, providing the overarching principles and framework for biological evaluation [18]. It guides manufacturers through the process of identifying, assessing, and managing biological risks based on the device's materials, design, and intended use [18]. The standard incorporates key concepts from ISO 14971, including hazard identification, risk estimation, risk evaluation, and risk control, specifically focused on biological harms [20]. The 2025 version of ISO 10993-1 further strengthens this risk-based approach and introduces new considerations such as reasonably foreseeable misuse, which requires manufacturers to consider how devices might be used outside their intended instructions [20].
Categorization of Medical Devices and Test Selection
The biological evaluation process begins with categorizing the medical device based on two primary factors: the nature and duration of body contact. This categorization drives the selection of appropriate biological effects that need evaluation [19].
Contact Duration Categories
ISO 10993-1 defines three distinct categories for contact duration, which are critical in determining the extent of testing required [22]:
| Duration Category | Definition | Examples |
|---|---|---|
| Limited Exposure | ≤ 24 hours | Single-use surgical instruments, injection needles |
| Prolonged Exposure | >24 hours to 30 days | Temporary implants, wound dressings |
| Long-term Exposure | > 30 days | Permanent implants (e.g., bone screws, pacemakers) |
The 2025 update to ISO 10993-1 provides further clarification on calculating exposure, introducing concepts of total exposure period (number of contact days between first and last use) and distinguishing between daily contact and intermittent contact [20]. Furthermore, it emphasizes that if a device contains chemicals known to bioaccumulate, the contact duration should be considered long-term unless otherwise justified [20].
Nature of Body Contact
The standard classifies devices based on the nature of body contact into three main categories [19]:
- Surface Devices: Contact intact skin, mucosal membranes, or breached/compromised surfaces.
- External Communicating Devices: Indirect contact with blood, tissues, bone, or dentin.
- Implant Devices: Direct contact with bone, tissue, or blood.
The combination of contact duration and nature of body contact determines which biological effects (e.g., cytotoxicity, sensitization, irritation) require evaluation. This relationship is detailed in Table A.1 of ISO 10993-1, which serves as a starting point for developing a biological evaluation plan [19].
The "Big Three" in Biocompatibility Testing
Among the various tests outlined in the ISO 10993 series, three assessments—cytotoxicity, sensitization, and irritation—are considered fundamental and are required for almost all medical devices, regardless of their categorization [21]. These are often referred to as the "Big Three" in biocompatibility testing.
Cytotoxicity Testing (ISO 10993-5)
Purpose: Cytotoxicity testing evaluates whether a medical device or its extracts have the potential to cause cell death or inhibit cell growth [21]. This test serves as an essential screening tool for identifying materials that may be toxic to living cells.
Experimental Protocols:
- Test System: Commonly uses mammalian cell lines such as Balb 3T3 (fibroblasts), L929 (fibroblasts), or Vero (kidney-derived epithelial cells) [21].
- Sample Preparation: Devices are typically tested as extracts prepared by immersing the device in extraction solvents such as physiological saline, vegetable oil, or cell culture medium under specified conditions [21]. ISO 10993-12 provides detailed guidance on sample preparation [16] [21].
- Exposure and Analysis: Cells are exposed to device extracts for approximately 24 hours, after which endpoints including cell viability, morphological changes, cell detachment, and cell lysis are assessed [21].
- Viability Assays: Quantitative methods include MTT, XTT, and neutral red uptake assays. Other methods such as Bradford protein, Chrystal violet, Resazurin dye, and Trypan blue assays are used less frequently [21].
Acceptance Criteria: While ISO 10993-5 does not define strict acceptance criteria, it provides guidance for data interpretation. Cell survival of 70% and above is generally considered a positive indicator, particularly when testing neat extract [21].
Sensitization Testing (ISO 10993-10)
Purpose: This assessment determines whether a medical device has the potential to cause allergic contact dermatitis through repeated or prolonged exposure to device extracts [16] [21].
Experimental Protocols:
- Test System: Traditionally conducted in vivo using guinea pig models (e.g., Guinea Pig Maximization Test or Buehler Test).
- Alternative Approaches: Increasingly, in vitro and in chemico methods are being developed and validated as alternatives to animal testing, aligning with the "3Rs" principles (Replacement, Reduction, and Refinement) [21].
- Sample Preparation: Device extracts are prepared using polar and non-polar solvents to ensure comprehensive extraction of potential sensitizers.
Irritation Testing (ISO 10993-23)
Purpose: Irritation testing evaluates the potential of a device, material, or their extracts to cause localized inflammatory responses (irritation) on skin, mucosal membranes, or other applicable tissues [16] [21].
Experimental Protocols:
- Test Systems: Can include in vitro reconstructed human tissue models (e.g., skin or epithelial models) or in vivo models.
- Sample Application: Extracts or materials are applied to the test system, and responses such as redness, swelling, or other signs of inflammation are evaluated.
- Scoring: Responses are typically scored using established grading systems to classify the irritation potential.
In Vitro vs. In Vivo Testing: A Comparative Analysis
The ISO 10993 standards incorporate both in vitro (conducted in controlled laboratory environments outside living organisms) and in vivo (conducted in or on living organisms) testing methodologies [2]. Each approach offers distinct advantages and limitations, which are particularly relevant in the context of biomaterial testing results comparison.
Comparative Analysis of Testing Methodologies
| Parameter | In Vitro Testing | In Vivo Testing |
|---|---|---|
| Definition | Tests performed outside living organisms (e.g., in petri dishes, test tubes) [2] | Tests performed within living organisms (animals or humans) [2] |
| Complexity | Lower complexity; reduced biological system interactions [2] [23] | Higher complexity; accounts for whole-organism interactions [2] |
| Control | High degree of environmental control [2] | Limited control due to biological variability [2] |
| Cost & Duration | Generally lower cost and shorter duration [24] | Higher cost and longer duration [2] |
| Ethical Considerations | Minimal ethical concerns [21] | Significant ethical considerations, especially for animal studies [21] |
| Regulatory Acceptance | Well-established for specific endpoints (e.g., cytotoxicity) [21] | Often required for more complex endpoints (e.g., implantation) [16] |
| Predictive Value | May not fully replicate conditions inside living organisms [2] [24] | Provides information on effects in whole, living organisms [2] |
| Standardization | Highly standardized for specific tests (e.g., ISO 10993-5 for cytotoxicity) [21] | Standardized protocols with defined animal welfare requirements (ISO 10993-2) [16] |
The Integrated Testing Approach
The ISO 10993 standards promote a sequential testing approach where in vitro studies often serve as foundational investigations to examine drug interactions and disease processes at the cellular level, while in vivo studies expand on this data by monitoring biological responses in living organisms [2]. According to ISO 10993-1, animal testing should only be conducted when existing scientific data and in vitro studies fail to provide adequate information for a comprehensive safety assessment [21]. This approach aligns with the "3Rs" principles (Replacement, Reduction, and Refinement) endorsed in Directive 2010/63/EU [21].
Experimental Workflows in Biological Evaluation
The biological evaluation of medical devices follows a structured, iterative process within a risk management framework. The following diagram illustrates the key stages of this workflow:
Biological Evaluation Workflow in ISO 10993
Test Selection Logic Based on Device Characteristics
The selection of specific tests required for a medical device depends on its categorization according to the nature of body contact and contact duration. The following diagram illustrates this decision-making process:
Test Selection Logic Based on Device Characteristics
Essential Research Reagents and Materials
Biocompatibility testing requires specific reagents, materials, and test systems to ensure reliable and reproducible results. The following table details key research reagent solutions used in ISO 10993-compliant testing:
| Reagent/Material | Function in Testing | Application Examples |
|---|---|---|
| Mammalian Cell Lines (L929, Balb/3T3) | Assess cell viability and morphological changes in response to device extracts [21] | Cytotoxicity testing (ISO 10993-5) |
| Extraction Solvents (Saline, DMSO, Vegetable Oil) | Extract leachable substances from device materials under standardized conditions [21] | Sample preparation for multiple test types |
| MTT/XTT/Neutral Red | Colorimetric indicators of cell viability and metabolic activity [21] | Quantitative cytotoxicity assessment |
| Guinea Pigs (Hartley, Dunkin-Hartley) | Traditional model for assessing sensitization potential [21] | Sensitization testing (in vivo) |
| Reconstructed Human Tissues (Skin, Epithelial models) | Alternative to animal testing for irritation and corrosion assessment [21] | In vitro irritation testing |
| Reference Materials (USP, negative/positive controls) | Benchmark for comparing test results and ensuring assay validity [16] [21] | Quality control across test systems |
| Culture Media & Supplements (RPMI, DMEM, FBS) | Support cell growth and maintenance during testing periods [21] | Cell-based assay systems |
The ISO 10993 series provides a comprehensive, risk-based framework for evaluating the biological safety of medical devices, balancing thorough safety assessment with ethical considerations regarding animal testing. The standards emphasize a scientifically rigorous approach that begins with material characterization and device categorization, followed by a tailored testing strategy based on the nature and duration of body contact. While the "Big Three" tests—cytotoxicity, sensitization, and irritation—form the foundation of biocompatibility assessment for nearly all devices, additional evaluations are required based on specific device characteristics and intended use.
The evolving landscape of medical device regulation, particularly the upcoming ISO 10993-1:2025 standard, continues to strengthen the integration of biological evaluation within a risk management framework and introduces important considerations such as reasonably foreseeable misuse. For researchers and drug development professionals, understanding the comparative advantages and limitations of in vitro versus in vivo testing methodologies remains crucial for designing efficient yet comprehensive biological evaluation plans that meet both scientific and regulatory requirements.
The Fundamental Strengths and Inherent Limitations of Each Approach
In biomedical research, in vitro (Latin for "in glass") and in vivo (Latin for "within the living") represent two fundamental experimental approaches with distinct philosophical and practical applications [10] [25]. In vitro methodologies involve testing biological components—such as cells, tissues, or organs—outside their natural biological context in controlled laboratory environments like petri dishes or test tubes [4]. These systems allow researchers to isolate specific variables and study biological processes in a simplified, reductionist manner. In contrast, in vivo methodologies involve experimentation within intact living organisms, preserving the complex physiological interactions between cells, tissues, and organ systems that characterize biological function [10]. This approach provides a holistic perspective that acknowledges the emergent properties of biological systems.
The choice between these methodologies represents a fundamental trade-off between experimental control and biological relevance. While in vitro systems offer precision and manipulation at the cost of physiological complexity, in vivo systems provide comprehensive biological context at the expense of experimental control [10] [25] [4]. This guide objectively examines the strengths and limitations of each approach within the context of biomaterial testing, providing researchers with evidence-based frameworks for methodological selection and interpretation of results.
Fundamental Principles and Definitions
In Vitro Testing Paradigm
The in vitro paradigm centers on studying biological components in isolation from their native environment. This approach typically utilizes cell cultures that fall into four main types: primary cultures (derived directly from tissues), secondary cultures (subcultures from primary ones), continuous cell lines (which can divide indefinitely), and hybridomas (used for monoclonal antibody production) [10]. Recent technological advances have expanded this paradigm to include more physiologically relevant models such as 3D tissue cultures, organ-on-a-chip systems (microfluidic devices that simulate key human organ functions), and microphysiological systems that recapitulate the function of entire organs in vitro [10] [4]. These systems combine tissue engineering, microfabrication, and biosensors to enable the organization and function of multiple tissue-specific cells over time, better approximating in vivo conditions while maintaining experimental control [4].
In Vivo Testing Paradigm
The in vivo paradigm maintains that meaningful understanding of biological processes requires studying them within the context of intact organisms, where complex interactions between different biological systems remain operational [10]. This approach utilizes whole living organisms, ranging from invertebrate models like Drosophila melanogaster (fruit fly) and Danio rerio (zebrafish) to vertebrate models including rodents (mice and rats) and, in specific cases, non-human primates [10]. These models allow observation of the effects of an intervention within the real context of a complete biological system, where the complex interactions between cells, tissues, and organs are preserved [10]. The in vivo approach is particularly valuable for assessing systemic effects, long-term toxicity, and complex pharmacokinetic profiles that emerge from the interplay of multiple biological systems [10] [26].
Comparative Analysis: Strengths and Limitations
Experimental Control and Complexity
The fundamental tension between experimental control and biological complexity represents the core trade-off when selecting between in vitro and in vivo approaches. In vitro systems enable researchers to isolate specific variables through stricter experimental control, reducing interference from systemic variables and improving result reproducibility compared to in vivo models [10]. This controlled environment allows precise manipulation of individual factors—such as pH, temperature, oxygen tension, and nutrient availability—that would be impossible to control in a living organism [10] [27]. However, this control comes at the cost of biological complexity, as in vitro systems cannot capture the inherent complexity of organ systems and the internal environment of the human body [25] [28].
In vivo approaches preserve this complexity, providing full physiological relevance by considering all metabolic processes and organic interactions [10]. The intact organism maintains the natural microarchitecture and compartmentation of organs, which enables compensatory mechanisms that may be lost in vitro [28]. For example, research on ammonia detoxification in liver models demonstrated that the organ's ability to switch metabolic pathways under stress conditions was extremely difficult to reproduce in vitro due to the loss of compartmentalization between periportal and pericentral regions of the liver lobule [28]. This preservation of biological complexity makes in vivo models indispensable for studying emergent properties that arise from system-level interactions.
Physiological Relevance and Human Applicability
The translation of research findings to human applications presents distinct challenges for both approaches. In vitro systems benefit from the ability to use human tissues obtained through surgery or donation, which enhances physiological relevance and human applicability [10]. Advanced systems like organ-on-a-chip technologies and 3D tissue models containing differentiated cell layers can physiologically represent complex tissues and evaluate specific endpoints, providing more human-relevant data than animal models in some contexts [10] [4]. However, these systems face significant limitations, as isolated and cultivated primary cells usually differ strongly from the corresponding cell type in an organism [28]. For example, when primary hepatocytes are isolated from their normal microenvironment, hundreds of genes are up- or down-regulated, potentially altering their response to experimental treatments [28].
In vivo models, particularly those using animal subjects, allow researchers to evaluate interventions within the context of an entire physiological system, including endocrine, nervous, and immune systems that interact in ways that cannot be fully replicated in vitro [10] [29]. However, the use of animal models introduces concerns about translatability due to physiological differences between species [25]. While mice share approximately 95% of their genes with humans, differences in drug absorption, distribution, metabolism, and excretion can limit the direct application of animal study results to human contexts [4].
Practical Considerations: Time, Cost, and Throughput
Practical considerations significantly influence methodological selection in research design. In vitro systems generally offer rapid results, are suitable for high-throughput testing, and are economically favorable compared to animal testing [25] [27]. The relative simplicity of these systems enables screening of large compound libraries efficiently, making them ideal for early-stage discovery research [10] [4]. Additionally, technological advances have increased the throughput of in vitro systems while reducing inter-experimental variability through standardized protocols and automated systems [30] [27].
In vivo studies are typically very expensive, require long and extensive timelines, and involve strict regulations and compliance standards due to their use of live subjects [25] [4]. The high costs derive from animal procurement, housing, veterinary care, and the specialized personnel required for animal handling [31]. Additionally, in vivo protocols require approval from Institutional Animal Care and Use Committees (IACUCs) and must adhere to evolving ethical standards for animal research, adding administrative burdens to these studies [31].
Table 1: Comprehensive Comparison of In Vitro and In Vivo Approaches
| Parameter | In Vitro | In Vivo |
|---|---|---|
| Experimental Control | High control over variables; reduced systemic interference [10] | Limited control due to complex biological interactions [10] |
| Biological Complexity | Limited; cannot replicate full organ system interactions [32] [25] | High; preserves complete physiological context [10] |
| Physiological Relevance | Enhanced when using human tissues, but cells may dedifferentiate [10] [28] | High for system-level responses, but species differences exist [25] [29] |
| Time Requirements | Relatively fast results [25] [4] | Long and extensive timelines [25] [4] |
| Cost Considerations | Relatively low cost [25] [4] | Very expensive [25] [4] |
| Throughput Capacity | Suitable for high-throughput screening [27] | Limited throughput due to practical constraints [31] |
| Regulatory Oversight | Less stringent than animal studies [25] | Strict regulations and compliance standards [25] [4] |
| Ethical Considerations | Reduced ethical concerns [10] [27] | Significant ethical implications, especially regarding animal use [10] [31] |
Technical Limitations and Methodological Constraints
Both approaches face distinct technical challenges that can impact data interpretation and application. In vitro systems struggle with difficulties in capturing interactions between different cell types, problems in extrapolating from in vivo doses to in vitro concentrations, and challenges in simulating the consequences of long-term exposures [32] [28]. These systems also face limitations in including xenobiotic metabolism into assays and extrapolating from perturbed pathways or biomarkers in vitro to adverse effects in vivo [28]. For example, in vitro skin models cannot recreate the full complexity of human skin, which contains a plethora of cell types including sensory neurons, tissue-resident immune cells, vascular cells, and lymphatic cells [32].
In vivo methodologies contend with higher interindividual variability, which can hinder interpretation in early research stages [10]. This variability stems from genetic differences, environmental factors, and complex biological interactions that are difficult to control [10] [26]. Additionally, in vivo models can produce confounding results due to systemic compensation mechanisms that may mask direct effects of an intervention [29]. For instance, in immune system research, the redundancy of signaling pathways in intact organisms can complicate the interpretation of gene knockout studies, as other pathways may compensate for the lost function [29].
Experimental Applications and Case Studies
Cytotoxicity Testing of Biomaterials
Cytotoxicity testing represents a critical application where both approaches provide complementary information. The ISO 10993-5 standard identifies three types of cytotoxicity tests: extract tests, direct contact tests, and indirect contact tests [30]. A recent study on Mg-1%Sn-2%HA composite for orthopedic implants demonstrated a standardized in vitro approach using L-929 mouse fibroblast cells cultured at 37°C with 5% CO₂ for 7 days [30]. The researchers used the elution technique in Dulbecco's Modified Eagle Medium (DMEM) supplemented with fetal bovine serum (FBS) to prepare extracts, which were then evaluated microscopically for aberrant cell morphology and degeneration, followed by quantitative cell toxicity assessment using the MTT method [30].
The results indicated high cell viability of 71.51% with the undiluted extract preparation, confirming the non-cytotoxic properties of the composite [30]. Furthermore, cell viability improved with dilution, attaining 84.93%, 93.20%, and 96.52% at concentrations of 50%, 25%, and 12.5%, respectively [30]. No notable morphological alterations or indications of cellular deterioration were observed [30]. This in vitro data provided preliminary safety evidence before proceeding to more complex and costly in vivo studies, demonstrating how in vitro approaches can serve as efficient screening tools in biomaterial development.
Drug Combination Studies in Oncology
Drug combination therapy represents another area where both approaches provide essential, complementary data. While in vitro models offer efficient platforms for initial synergy screening, in vivo models better capture tumor heterogeneity and mimic tumor molecular features and treatment responses [26]. However, in vivo models often demonstrate substantial heterogeneity in treatment effects among animals, which, while closely reflecting clinical outcomes, poses challenges for accurate evaluation of treatment responses [26].
The SynergyLMM framework was developed specifically to address the statistical challenges of in vivo drug combination studies [26]. This comprehensive modeling approach accommodates complex experimental designs, including multi-drug combinations, and offers practical options for statistical analysis of both synergy and antagonism through longitudinal drug interaction analysis [26]. The method supports the use of various synergy scoring models, such as Bliss independence, Highest Single Agent (HSA), and Response Additivity (RA), accompanied by uncertainty quantification and statistical assessment [26].
In a reanalysis of drug combination experiments from Narayan et al., SynergyLMM revealed important nuances that simple endpoint analyses missed [26]. For the U87-MG Fluc-Mcherry orthotopic glioblastoma model with Docetaxel + GNE-317 combination, the analysis showed significant synergy under the HSA model but not enough statistical evidence to reject additivity under the Bliss model [26]. This contrasted with the original authors' conclusions using the median-effect principle, demonstrating how methodological choices can significantly impact interpretation of in vivo results.
Table 2: Experimental Considerations for In Vivo Drug Combination Studies
| Consideration | Challenge | SynergyLMM Solution |
|---|---|---|
| Longitudinal Data | Traditional endpoint analyses lose temporal information [26] | Uses all longitudinal measurements to capture dynamic effects [26] |
| Reference Models | Different models (Bliss, HSA, RA) can yield conflicting results [26] | Supports multiple reference models to assess conclusion robustness [26] |
| Inter-animal Variability | High heterogeneity complicates effect detection [26] | Uses mixed effects models to account for inter-animal variation [26] |
| Experimental Design | Determining appropriate animal numbers and measurement frequency [26] | Provides power analysis tools to optimize study design [26] |
| Model Selection | Choosing appropriate tumor growth kinetics model [26] | Supports both exponential and Gompertz growth models with diagnostics [26] |
Immunological Applications
Immunological research particularly highlights the complementary strengths of both approaches. In vitro models are invaluable for reducing the complexity of the immune system to manageable components, allowing detailed study of specific signaling pathways and mediator release [29]. For example, in vitro systems have been essential for delineating the molecular mechanisms of Toll-like receptor recognition and the cytolytic machinery of natural killer cells and cytotoxic T lymphocytes [29]. These reductionist approaches enable researchers to isolate specific immune functions without the confounding effects of entire system integration.
However, the immune system fundamentally operates through complex cellular interactions and spatial organizations that are difficult to reconstruct in vitro [29]. The transition of immune cells from blood vessels to tissues, for instance, involves a coordinated multi-step process (rolling, activation, firm adhesion, and transmigration) that depends on the vascular architecture and flow conditions not easily replicated in vitro [29]. Similarly, the compartmentalization of immune responses between primary and secondary lymphoid organs represents a system-level property that requires intact organisms for meaningful study [29]. Therefore, while in vitro systems provide mechanistic insights, in vivo models remain essential for understanding immune function in physiologically relevant contexts.
Experimental Design and Methodological Protocols
Standardized In Vitro Cytotoxicity Testing
For biomaterial cytotoxicity assessment, researchers should follow established standards such as ISO 10993-5, which provides frameworks for test selection and implementation [30]. A typical protocol involves:
Sample Preparation: Using the elution method to prepare extracts in culture medium supplemented with serum, with incubation typically ranging from 24-72 hours at 37°C [30].
Cell Culture: Maintaining appropriate mammalian cells (e.g., L-929 mouse fibroblast cells) under standard conditions (37°C, 5% CO₂) for a specified period, typically 7 days [30].
Assessment Methods:
- Microscopic Evaluation: Examining cell monolayers for aberrant morphology and degeneration [30].
- Quantitative Assessment: Using methods like MTT assay, which measures mitochondrial dehydrogenase activity by converting yellow MTT to purple formazan crystals, with absorbance measurement at 492 nm [30].
- Alternative Assays: Including colorimetric (MTS, XTT, LDH), fluorometric (CFDA-AM, alamar blue), and luminometric (ATP assay) approaches depending on research needs [30].
Interpretation: Comparing results to appropriate controls and established benchmarks for cytotoxicity classification [30].
This standardized approach ensures reproducibility and facilitates comparison across different studies and laboratories, providing reliable preliminary safety data for biomaterial development.
Advanced In Vivo Combination Therapy Studies
For in vivo drug combination studies, proper experimental design and statistical analysis are crucial for robust conclusions. The SynergyLMM framework provides a comprehensive workflow:
Input Data Collection: Longitudinal tumor burden measurements (e.g., volume, luminescence) in various treatment groups and control animals, normalized against treatment initiation time points [26].
Model Selection: Choosing between exponential or Gompertz tumor growth kinetics based on model diagnostics and performance metrics [26].
Statistical Modeling: Applying linear mixed effects models (exponential growth) or non-linear mixed effects models (Gompertz growth) to estimate growth parameters for each treatment group [26].
Model Diagnostics: Checking model assumptions, identifying outlier observations, and assessing influential subjects [26].
Synergy Assessment: Calculating time-resolved synergy scores using appropriate reference models (Bliss, HSA, or RA) with uncertainty quantification and statistical significance evaluation [26].
Power Analysis: Conducting post-hoc power analysis of completed experiments or a priori power analysis for new experiments to determine appropriate sample sizes and measurement frequencies [26].
This comprehensive approach addresses the specific challenges of in vivo combination studies, particularly the longitudinal nature of tumor growth measurements and substantial inter-animal heterogeneity [26].
Diagram 1: Methodological Selection Framework for Biomaterial Testing. This decision pathway illustrates the fundamental trade-offs between experimental control and biological complexity when selecting between in vitro and in vivo approaches.
Essential Research Reagent Solutions
Successful implementation of either methodology requires appropriate selection of research reagents and materials. The following table details essential components for representative experiments in both approaches:
Table 3: Essential Research Reagent Solutions for In Vitro and In Vivo Studies
| Reagent/Material | Application | Function | Representative Examples |
|---|---|---|---|
| Cell Culture Media | In vitro systems | Provide essential nutrients for cell growth and maintenance | Dulbecco's Modified Eagle Medium (DMEM) with fetal bovine serum (FBS) [30] |
| Biomaterial Extracts | In vitro cytotoxicity | Evaluate leachable substances from test materials | Mg-1%Sn-2%HA composite extracts in culture medium [30] |
| Viability Assays | In vitro assessment | Quantify cell health and metabolic activity | MTT assay, ATP assays, fluorometric assays [30] |
| Animal Models | In vivo studies | Provide complete biological context for testing | Rodent models, zebrafish, non-human primates [10] |
| Tumor Measurement Tools | In vivo oncology | Monitor disease progression and treatment response | Caliper measurements, luminescence imaging [26] |
| Statistical Frameworks | In vivo data analysis | Analyze complex longitudinal data and synergy | SynergyLMM for drug combination studies [26] |
The fundamental strengths and inherent limitations of in vitro and in vivo approaches underscore their essential complementarity in biomaterial testing. Rather than viewing these methodologies as mutually exclusive, researchers should recognize their synergistic potential when strategically integrated within a structured research program [10]. In vitro systems provide unparalleled efficiency, control, and mechanistic insights for early-stage screening and hypothesis generation, while in vivo models deliver indispensable physiological context and system-level validation for promising candidates [10] [25] [4].
This complementary relationship is reflected in evolving regulatory frameworks that acknowledge the value of both approaches. The European Union's REACH regulation prioritizes alternative methods over animal testing in the chemical sector, while the FDA and other agencies have begun accepting data generated through advanced in vitro platforms in pharmacology and toxicology contexts [10]. Similarly, the role of EURL ECVAM (European Union Reference Laboratory for Alternatives to Animal Testing) has been crucial in validating, harmonizing, and promoting in vitro and alternative studies while recognizing the current necessity of in vivo approaches for complex endpoints [10].
The future of biomaterial testing lies not in exclusive reliance on one approach, but in the continued development of more physiologically relevant in vitro systems and more refined, humanized in vivo models that collectively enhance predictive accuracy while addressing ethical concerns. Through strategic integration of both methodologies, researchers can maximize scientific rigor while advancing the fundamental goals of replacement, reduction, and refinement of animal use in research—the essential 3Rs principle that guides ethical scientific progress [10] [27].
The implantation of a biomaterial initiates a complex and predictable sequence of biological events known as the biological response continuum. This process begins with acute inflammation and progresses through chronic inflammation, granulation tissue development, and ultimately culminates in fibrous encapsulation—a process scientifically termed the foreign body response (FBR) [33]. Understanding this continuum is crucial for researchers and drug development professionals who must navigate the challenging transition from in vitro analysis to in vivo performance. The FBR represents a significant clinical challenge as it can compromise the functionality and long-term performance of implanted biomaterials, potentially leading to implant failure and compromised tissue regeneration [33]. This biological cascade occurs consistently across diverse biomaterial applications, including medical implants, drug delivery systems, and tissue engineering scaffolds, making its understanding fundamental to biomaterial research and development.
The foreign body response is not a single event but rather a synergistically destructive process involving proinflammatory macrophages and activated fibroblasts that drive uncontrolled inflammation and excessive fibrosis [33]. This complex interplay between the immune system and the implanted material determines the ultimate fate of any medical device introduced into the biological environment. The clinical significance of this process cannot be overstated, as the chronic damage resulting from implant rejection often outweighs the intended healing benefits, presenting a considerable challenge when implementing treatment-based biomaterials [33]. Consequently, comprehensive understanding of the biological response continuum is essential for developing safer and more effective biomaterials that can successfully integrate with host tissues.
The Foreign Body Response: A Stage-Based Paradigm
Phase Progression and Key Cellular Players
The foreign body response follows a well-defined sequence of biological events that occur when a biomaterial is introduced into living tissue. While the exact timing may vary based on material properties and implantation site, the fundamental progression remains consistent across most biomaterial applications [33]. The process begins immediately upon implantation and can continue for the duration of the material's presence in the body.
Table 1: Stages of the Foreign Body Response
| Stage | Time Frame | Key Cellular Players | Primary Molecular Mediators |
|---|---|---|---|
| Protein Adsorption | Immediate (seconds to minutes) | Plasma proteins (fibrinogen, albumin, immunoglobulins) | Complement proteins, coagulation factors |
| Acute Inflammation | 0-48 hours | Neutrophils | Reactive oxygen species (ROS), antimicrobial peptides, matrix metalloproteinases (MMPs) |
| Chronic Inflammation/Macrophage Activity | 48 hours onward | Macrophages, monocytes | TNF-α, IL-1β, IL-6, IL-8, IL-4, IL-10, IL-13 |
| Foreign Body Giant Cell Formation | Days to weeks | Multinucleated giant cells (fused macrophages) | Acid hydrolases, proteases, reactive oxygen species |
| Fibrous Capsule Formation | Weeks to months | Fibroblasts, myofibroblasts | TGF-β, PDGF, VEGF, collagen deposits |
Following protein adsorption, neutrophils swiftly dominate the acute inflammatory phase, arriving at the implantation site within hours [33]. These innate immune cells demonstrate potent phagocytic capabilities, engulfing foreign particles, debris, and potential pathogens associated with the biomaterial. Neutrophils also release an array of antimicrobial molecules, including reactive oxygen species (ROS) and antimicrobial peptides, which effectively eradicate potential threats and ensure tissue sterility [33]. Beyond their defensive role, neutrophils secrete various growth factors and matrix metalloproteinases (MMPs) that initiate extracellular matrix (ECM) remodeling—a crucial process for subsequent tissue integration or repair [33].
Within approximately 48 hours, a cellular transition occurs as neutrophils are gradually replaced by macrophages, marking the shift from acute inflammation to the chronic foreign body response [33]. Macrophages are pivotal regulators of this phase, possessing the ability to continuously proliferate and release chemotactic factors that recruit additional immune cells to the implantation site. These cells exert sophisticated control over the immune response through polarization—adopting distinct functional phenotypes based on local environmental cues [33]. The classically activated M1 macrophages exhibit proinflammatory properties and drive the FBR through the release of tumor necrosis factor-alpha (TNF-α), interleukin (IL)-1β, IL-6, and IL-8 [33]. In contrast, alternatively activated M2 macrophages coordinate anti-inflammatory responses and tissue remodeling through the secretion of IL-13, IL-10, and IL-4 [33].
When macrophages encounter biomaterials that resist phagocytosis due to size or structural properties, they may fuse together to form foreign body giant cells (FBGCs) [33]. These large multinucleated cells adhere persistently to biomaterial surfaces and release degradative enzymes including acid hydrolases, proteases, and reactive oxygen species. This enzymatic activity can lead to chronic inflammation and potentially compromise biomaterial functionality through surface degradation [33]. The presence of FBGCs represents a hallmark of the foreign body response and serves as a key indicator in histopathological evaluations of biomaterial compatibility.
The final phase of the biological response continuum involves the development of fibrous encapsulation, where fibroblasts are recruited to the implantation site and activated to deposit collagenous matrix [33]. This process is driven by growth factors released by macrophages, platelets, and adipocytes in the wound environment, particularly platelet-derived growth factors (PDGF), transforming growth factor β (TGF-β), and vascular endothelial growth factors (VEGF) [33]. The resulting fibrous capsule effectively walls off the implant from surrounding tissues, a biological attempt to isolate the foreign material that often compromises the intended function of medical devices and tissue engineering constructs.
Figure 1: The Foreign Body Response Cascade. This diagram illustrates the sequential biological events following biomaterial implantation, from initial protein adsorption to final fibrous encapsulation, highlighting key cellular players at each stage.
Comparative Experimental Models: In Vitro vs. In Vivo Assessment
The Translational Challenge
A critical challenge in biomaterial development lies in the significant disconnect between in vitro assessments and in vivo outcomes. A comprehensive multicenter review from the European consortium BioDesign, which analyzed data from 36 in vivo studies and 47 in vitro assays testing 93 different biomaterials, revealed a "surprisingly poor correlation between in vitro and in vivo testing of biomaterials for bone regeneration" [34]. The study found no significant overall correlation between in vitro and in vivo outcomes, with mean in vitro scores showing only a 58% trend of covariance to in vivo scores [34]. This substantial discrepancy highlights the inadequacies of current in vitro assessment methods and underscores the need for novel approaches to in vitro biomaterial testing and validated preclinical pipelines.
The biological complexity of in vivo environments introduces variables that are exceptionally difficult to replicate in laboratory settings. The immune response sensitivity differs significantly between connective tissue and bone tissue, with connective tissue generally responding more sensitively to foreign bodies such as bone substitute materials [35]. This was demonstrated in a comparative study of a novel hybrid bone substitute material, where subcutaneous implants elicited stronger inflammatory reactions with higher counts of polymorphonuclear cells, lymphocytes, multinucleated giant cells, and plasma cells at day 10, alongside consistently elevated irritancy scores compared to calvaria implants [35]. In contrast, calvaria implants showed increased neovascularization, reflecting bone-specific regenerative processes [35]. These findings underscore how implantation site dramatically influences immune activation, vascularization, and biomaterial resorption.
Standardized Methodologies for Biocompatibility Assessment
To ensure consistent evaluation across studies, researchers rely on standardized protocols for assessing biocompatibility and host tissue responses. The DIN EN ISO 10993-6 standard provides a critical framework for the macroscopic and microscopic evaluation of tissue responses to implanted materials, specifically addressing local effects after implantation [35]. This standard employs histopathological analysis to assess parameters such as inflammation, fibrosis, neovascularization, and bone remodeling through semi-quantitative scoring of cellular and tissue reactions surrounding the implant [35]. Histological sections are typically examined and scored on a 0 to 4 scale for various cell types (e.g., polymorphonuclear cells, lymphocytes, macrophages, giant cells) and tissue changes (e.g., necrosis, fibrosis, neovascularization) [35]. These scores calculate a total irritation or tissue response score, which helps classify a material's biocompatibility as non-irritant, slightly irritant, moderately irritant, or severely irritant.
For regulatory approval of bone substitute materials—particularly in the EU under Medical Device Regulation (MDR) or in the US under FDA guidance—both subcutaneous and calvarial implantation models in rats are frequently used in a stepwise preclinical testing strategy [35]. Initial biocompatibility and safety are typically assessed via subcutaneous implantation, while functional evaluation in bone tissue is tested using calvarial defect models or other load-bearing defect models, depending on the intended application [35]. Data from these models form essential components of the technical documentation submitted for regulatory review, demonstrating that materials are safe, biocompatible, and effective in supporting bone regeneration.
Table 2: Comparative Analysis of Implantation Models Based on ISO 10993-6 Standards
| Assessment Parameter | Subcutaneous Model | Calvarial Defect Model | Statistical Significance |
|---|---|---|---|
| Polymorphonuclear Cells | Higher counts at day 10 | Lower counts at day 10 | p≤0.05* |
| Lymphocytes | Elevated throughout study | Reduced presence | p≤0.05* |
| Plasma Cells | Moderate infiltration (decreasing over time) | Absent to minimal | p≤0.01 |
| Multinucleated Giant Cells | Consistently higher numbers | Fewer giant cells | p≤0.05* |
| Neovascularization | Lower vascularization | Significantly increased | p≤0.01 |
| Material Degradation | Significantly greater at day 60 | Slower degradation profile | p≤0.001* |
| Fibrosis | Minimal at early time points | Increased at day 60 | p≤0.05* |
| Bone Regeneration | Not applicable | Gradual, continuous increase | N/A |
Advanced Biomaterial Strategies to Modulate the Host Response
Material Property Engineering
Contemporary biomaterial research has evolved beyond simply minimizing immune recognition to actively engineering materials that modulate host responses. Key material properties—including surface chemistry, topography, and composition—have received significant attention for their ability to influence the foreign body reaction [33]. For instance, surface texture has been demonstrated to directly impact fibrotic encapsulation thickness, as evidenced by research showing that breast implants with specific surface characteristics affected the thickness of the surrounding fibrotic capsule [33]. This finding highlights how material design choices can directly influence clinical outcomes through modulation of the biological response continuum.
Decellularized scaffolds represent another promising approach, particularly those derived from placental membranes such as amniotic and chorionic tissues [36]. These materials leverage natural extracellular matrix compositions while eliminating cellular components that might trigger immune reactions. The human amniotic membrane (AM) demonstrates highly attractive biological properties, including low immunogenicity and anti-inflammatory, anti-fibrotic, anti-microbial, pain-relieving, and modulatory angiogenic effects [36]. When associated with chorion to create an amnion/chorion membrane (ACM), the resulting scaffold shows improved mechanical properties with a thirty-fivefold increase in thickness and more than tenfold increase in tearing and compression strength compared to decellularized AM alone [36]. These enhanced physical properties are achieved while maintaining biocompatibility and osteogenic potential, demonstrating how material combinations can optimize both biological and mechanical performance.
Immunomodulatory Design Approaches
The emerging frontier in biomaterial development involves creating immunomodulatory biomaterials that actively shape the immune environment to favor integration over rejection. One innovative strategy employs microporous annealed particle (MAP) scaffolds composed of hydrogel microparticles with inter- and intra-particle cross-links that provide structure and cell-scale porosity [37]. These injectable scaffolds have demonstrated remarkable ability to mitigate the foreign body response, with studies showing "no fibrous capsule or significant FBR was observed" even at 12 months post-implantation [37]. This represents a substantial improvement over traditional materials that typically provoke substantial foreign body reactions.
Further refinement of this approach incorporates bioactive components such as heparin microislands within MAP scaffolds, which leverage endogenous growth factor sequestration to direct cellular responses [37]. These heparin μislands "facilitate increased cell infiltration and recruit a wider variety of cells at 1 month than blank MAP scaffolds," though researchers note this effect diminishes after 3 months [37]. The strategic inclusion of bioactive molecules represents a shift from passive to active biomaterial design, where implants don't merely avoid provoking negative responses but actively promote regenerative processes. Long-term studies of these systems have revealed a potential "activation of the complement-mediated immune response at 12 months in both groups, possibly associated with pore collapse in the implants," highlighting the importance of evaluating biomaterial performance across extended timeframes [37].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagents and Materials for Biomaterial Evaluation
| Reagent/Material | Function in Research | Experimental Example |
|---|---|---|
| Biphasic Hydroxyapatite/β-TCP Granules | Osteoconductive scaffold providing controlled resorption | maxresorb flexbone hybrid bone substitute [35] |
| Type I Collagen Matrix | Mimics natural ECM, promotes cell attachment | Porcine collagen matrix in hybrid bone blocks [35] |
| Decellularized Amnion/Chorion Membrane | Biocompatible scaffold with improved mechanical properties | DL-ACM for guided bone regeneration [36] |
| Heparin μIslands | Growth factor sequestration to enhance bioactivity | Modified heparin in MAP scaffolds for accelerated tissue formation [37] |
| PEG-based Hydrogels | Tunable scaffold material with controllable physical properties | Microporous annealed particle (MAP) scaffolds [37] |
| Technovit 9100 | Plastic embedding medium for histological processing | Tissue processing for undecalcified bone samples [35] |
| Histological Staining Reagents | Tissue structure visualization and cellular identification | H&E staining for general histology; Masson's trichrome for collagen [35] [33] |
Figure 2: Comprehensive Workflow for Biomaterial Evaluation. This diagram outlines the standardized pathway from initial material development through regulatory submission, highlighting key decision points and methodological requirements at each stage.
The biological response continuum—from acute inflammation to fibrous encapsulation—represents a fundamental challenge in biomaterial science and regenerative medicine. Understanding this sequential process is essential for developing advanced materials that can successfully integrate with host tissues rather than provoking rejection. The significant discrepancy between in vitro predictions and in vivo outcomes underscores the complexity of biological systems and the limitations of current laboratory models [34]. This disconnect necessitates a combined testing approach utilizing both subcutaneous and site-specific implantation models to fully characterize material performance [35].
Future directions in biomaterial development point toward actively immunomodulatory designs that don't merely avoid immune recognition but strategically direct host responses toward regenerative outcomes [33] [37]. The continued refinement of decellularized scaffolds [36], advanced hydrogel systems [37], and strategically bioactive materials represents promising pathways to better clinical outcomes. As research advances, the focus must remain on comprehensive evaluation across extended timeframes and in biologically relevant models to ensure that promising in vitro results translate to successful in vivo performance and ultimately, improved patient outcomes.
A Practical Guide to Test Methods and Their Strategic Application
In the landscape of biomaterials research and drug development, the journey from laboratory concept to clinical application is paved with rigorous safety assessments. In vitro assays provide a critical first line of defense, enabling researchers to evaluate biological responses in controlled, reproducible environments outside of living organisms (in glass) [38]. These preliminary screenings are indispensable within the broader thesis of comparing in vitro and in vivo (within the living) test results, as they offer a cost-effective, ethically favorable, and highly controlled means of identifying potential safety concerns before committing to complex and costly animal studies [38] [10]. Among the essential suite of tests, three assays stand as fundamental pillars: cytotoxicity, which assesses cell death and metabolic impairment; hemocompatibility, which evaluates interactions with blood components; and genotoxicity, which identifies damage to genetic material [39] [30]. This guide provides a comparative analysis of these assays, detailing experimental protocols, key reagents, and data interpretation to inform the work of researchers and drug development professionals.
Experimental Protocols for Key In Vitro Assays
Standardized and reliable experimental protocols are the foundation of reproducible and meaningful in vitro safety data. The following section outlines established methodologies for these three critical assays, drawing from international standards and recent peer-reviewed research.
Cytotoxicity Assay (MTT Method)
The MTT assay is a widely used colorimetric method for measuring cellular metabolic activity as an indicator of cell viability, proliferation, and cytotoxicity [30]. It is performed per ISO 10993-5 guidelines [30].
- Objective: To evaluate the cytotoxic potential of a Mg-1%Sn-2%HA composite by assessing the viability of L-929 mouse fibroblast cells after exposure to the material's extracts.
- Cell Culture: L-929 mouse fibroblast cells are cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with fetal bovine serum (FBS) at 37°C in a 5% CO₂ atmosphere [30].
- Extract Preparation: The test material is extracted using the elution method in the complete cell culture medium (DMEM with FBS) for a specified duration [30].
- Exposure and Incubation: The cultured cell monolayers are exposed to the prepared extract (100% concentration) and a series of dilutions (e.g., 50%, 25%, 12.5%). The cells are then incubated for a predetermined period, typically 7 days [30].
- Viability Measurement: After incubation, the MTT reagent is added. Mitochondrial dehydrogenases in viable cells convert the yellow MTT tetrazolium salt into purple formazan crystals. These crystals are dissolved using an organic solvent like Dimethyl Sulfoxide (DMSO), and the solution's absorbance is measured at 492 nm [30]. The absorbance is directly proportional to the number of viable cells.
- Data Analysis: Cell viability is calculated as a percentage relative to the untreated control group. A cell viability of 71.51% with an undiluted extract, as seen in a study on a Mg-based composite, indicates non-cytotoxicity according to ISO standards, with viability typically increasing with higher extract dilutions [30].
Hemocompatibility Assay
Hemocompatibility testing assesses the adverse effects of a biomaterial on blood, such as hemolysis (the destruction of red blood cells) [39].
- Objective: To determine the hemolytic potential of silk fibroin nanoparticles (SFN) through in vitro blood incubation.
- Blood Preparation: Fresh human blood is collected with an anticoagulant, such as sodium citrate, and red blood cells (RBCs) are isolated by centrifugation and washing with phosphate-buffered saline (PBS) [39].
- Test Incubation: The test material (e.g., SFN) is incubated with the prepared RBC suspension under controlled conditions (e.g., 37°C for a set time) [39].
- Positive and Negative Controls: A positive control (e.g., distilled water causing complete hemolysis) and a negative control (e.g., PBS causing minimal hemolysis) are run in parallel.
- Measurement: After incubation, the mixture is centrifuged. The release of hemoglobin into the supernatant due to RBC damage is measured spectrophotometrically at a wavelength of 540 nm [39].
- Data Analysis: The percentage of hemolysis is calculated by comparing the absorbance of the test sample to the positive and negative controls. A low hemolysis percentage indicates good hemocompatibility. Studies on dual-labeled silk fibroin nanoparticles have shown only a mild host response in blood, suggesting favorable hemocompatibility [39].
Genotoxicity Assay (Cytokinesis-Block Micronucleus - CBMN)
The CBMN assay is a comprehensive method for detecting genotoxic effects by measuring the formation of micronuclei in the cytoplasm of interphase cells, which originate from chromosome fragments or whole chromosomes lagging during cell division [39].
- Objective: To evaluate the genotoxic potential of [99mTc]Tc-FITC-Silk Fibroin Nanoparticles.
- Cell Culture: Human lymphocytes or other mammalian cell lines are cultured in vitro.
- Cell Exposure and Cytokinesis Block: Cells are exposed to the test material. Cytochalasin B is then added to the culture. Cytochalasin B inhibits cytokinesis—the division of the cytoplasm—while allowing nuclear division to proceed. This results in the formation of binucleated cells.
- Cell Harvesting and Staining: After a specified incubation period, cells are harvested, placed on slides, and stained with a DNA-specific stain (e.g., Giemsa, DAPI).
- Microscopic Analysis: Binucleated cells are scored under a microscope for the presence of one or more micronuclei. A micronucleus is a small, extranuclear body that represents genetic material that was not incorporated into the daughter nuclei during cell division.
- Data Analysis: The frequency of micronucleated binucleated cells is compared between treated and control cultures. A statistically significant increase in frequency indicates genotoxic potential. For instance, the CBMN assay applied to radiolabeled silk fibroin nanoparticles did not show induced genotoxicity under the described test conditions [39].
Comparative Analysis of Assay Data
To facilitate a clear comparison of outcomes and sensitivity across the three assays, quantitative data from research studies is summarized in the table below.
Table 1: Comparative Quantitative Data from In Vitro Safety Assays
| Assay Type | Test Material | Key Metric | Result | Interpretation |
|---|---|---|---|---|
| Cytotoxicity (MTT) | Mg-1%Sn-2%HA composite [30] | Cell Viability (%) | 71.51% (undiluted), 96.52% (12.5% dilution) | Non-cytotoxic (ISO 10993-5) |
| Hemocompatibility | Silk Fibroin Nanoparticles (SFN) [39] | Hemolysis (%) | Low, mild host response | Hemocompatible |
| Genotoxicity (CBMN) | [99mTc]Tc-FITC-SFN [39] | Micronucleus Frequency | No significant increase | Non-genotoxic |
The Scientist's Toolkit: Essential Research Reagents and Materials
The execution of these standardized assays requires a consistent set of high-quality reagents and instruments. The following table details the core components of the research toolkit for these in vitro evaluations.
Table 2: Essential Research Reagent Solutions for In Vitro Safety Assays
| Item | Function / Application | Examples / Specifications |
|---|---|---|
| L-929 Mouse Fibroblast Cells | A standard cell line used for cytotoxicity testing per ISO 10993-5 [30]. | Mammalian cell culture. |
| DMEM with FBS | Cell culture medium; provides nutrients and growth factors for cell maintenance and growth during testing [30]. | Dulbecco's Modified Eagle Medium supplemented with Fetal Bovine Serum. |
| MTT Reagent | A yellow tetrazolium salt used in cytotoxicity assays; reduced to purple formazan by metabolically active cells [30]. | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide. |
| Dimethyl Sulfoxide (DMSO) | An organic solvent used to dissolve the insoluble purple formazan crystals produced in the MTT assay for spectrophotometric reading [30]. | |
| Human Red Blood Cells (RBCs) | Used directly in hemocompatibility assays to test the hemolytic potential of a material upon contact with blood [39]. | Isolated from fresh whole blood. |
| Phosphate Buffered Saline (PBS) | A balanced salt solution used as a negative control in hemolysis assays and for washing cells [39]. | |
| Cytochalasin B | A reagent used in the CBMN genotoxicity assay to inhibit cytokinesis, resulting in binucleated cells for accurate micronucleus scoring [39]. | |
| Spectrophotometer / Microplate Reader | An instrument used to measure the absorbance of solutions in colorimetric assays like MTT and hemolysis [30]. |
Cytotoxicity, hemocompatibility, and genotoxicity assays form an indispensable triad in the preliminary safety assessment of biomaterials and pharmaceuticals. The experimental data and protocols outlined here provide a framework for objective comparison and validation of product safety. While these in vitro methods are powerful tools for screening and mechanistic studies, they represent an initial phase within a more comprehensive testing strategy. As dictated by the broader thesis of in vitro versus in vivo comparison, data from these controlled systems must be interpreted with an understanding of their limitations in modeling the full complexity of a living organism [38] [10]. The trend in regulatory frameworks is increasingly toward accepting advanced in vitro models, yet a complete safety profile often still requires complementary in vivo studies to confirm biocompatibility and efficacy in a whole-body context, ensuring that promising laboratory findings can be safely and effectively translated into clinical applications [10].
In the field of biomaterial and medical device development, in vivo evaluations are indispensable for assessing biological safety and functional performance within a living organism. These studies provide critical data on complex whole-body interactions that cannot be fully replicated in laboratory settings. Sensitization, irritation, and implantation testing represent three core in vivo evaluation categories required by international standards such as ISO 10993 for biological safety assessment. These tests collectively determine how the human body responds to materials intended for medical use, identifying potential adverse effects like allergic reactions, inflammatory responses, and long-term tissue compatibility issues. While in vitro methods offer valuable preliminary data, in vivo studies remain the gold standard for predicting clinical performance and ensuring patient safety, particularly for devices with prolonged or permanent tissue contact.
Comparative Analysis of In Vivo and In Vitro Testing Methods
Table 1: Comparison of In Vivo and In Vitro Approaches for Core Biological Evaluations
| Evaluation Type | In Vivo Models | In Vitro Alternatives | Key Advantages of In Vivo | Key Limitations of In Vivo |
|---|---|---|---|---|
| Sensitization | Guinea Pig Maximization Test (GPMT), Buehler Test, Murine Local Lymph Node Assay (LLNA) [40] [41] | Direct Peptide Reactivity Assay, Human Cell Line Activation Test (h-CLAT), KeratinoSens [42] [41] | Models complete immune system response including neuro-immune interactions; Essential for detecting delayed hypersensitivity [40] [43] | Ethical concerns; Costly and time-consuming; Subject to interspecies variability [40] [43] |
| Irritation | Rabbit Skin Irritation Test, Intracutaneous Reactivity Test [40] | Reconstructed human epidermis (RhE) models, Full-thickness human skin equivalents (HSE) [42] | Captures complex inflammatory cascades and vascular responses [40] | Ethical considerations; Difficult to quantify subjective symptoms like burning or itching [42] |
| Implantation | Rat, mouse, rabbit, or canine models with tissue implantation for 1-12+ weeks [44] [7] | Macrophage-biomaterial interaction assays, 3D tissue cultures [7] | Assesses dynamic tissue integration, foreign body reaction, and long-term degradation in situ [44] [7] | High cost; Extended duration; Complex surgical procedures; Ethical regulations [44] [7] |
Table 2: Quantitative Data from Representative In Vivo Studies
| Study Focus | Model System | Key In Vivo Endpoints | Results | Source |
|---|---|---|---|---|
| Magnesium Composite Implantation | Rat femoral defect model (3 months) [44] | Hydrogen gas evolution, fibrotic response, new bone formation [44] | No or minimal gas evolution and fibrotic response with osteointegration [44] | Frontiers in Bioengineering and Biotechnology (2025) [44] |
| Host Response to Biomaterials | Rat abdominal wall defect (14, 35 days) [7] | Remodeling score (0-3 scale: foreign body reaction, degradation, tissue organization) [7] | ECM materials: higher remodeling scores; Synthetic materials: lower scores indicating foreign body reaction [7] | Tissue Engineering Part C (2014) [7] |
| Sensitization Reference Material | Guinea Pig Maximization Test [41] | Positive sensitization response [41] | DNCB-PU sheet extracts elicited positive responses [41] | Journal of Biomedical Materials Research (2024) [41] |
Sensitization Testing: Methodologies and Protocols
Experimental Protocols for In Vivo Sensitization Assessment
Sensitization testing evaluates the potential of a material or device to cause allergic contact dermatitis, a delayed-type hypersensitivity reaction. The Guinea Pig Maximization Test (GPMT) is a extensively validated in vivo method. The protocol involves an induction phase where the test material extract or suspension is intradermally injected with Freund's Complete Adjuvant to enhance immune response, followed by a topical challenge phase after a 10-14 day rest period. Reactions are graded based on erythema and edema formation at challenge sites compared to controls [40] [41]. The Local Lymph Node Assay (LLNA) represents a refined in vivo alternative that reduces animal suffering by measuring lymphocyte proliferation in draining lymph nodes following topical application, without the use of adjuvants. This assay provides a quantitative endpoint through incorporation of radioactive thymidine or alternative markers like bromodeoxyuridine, with a stimulation index ≥3 considered positive [40] [41].
Correlation with Human Response and Testing Considerations
While animal sensitization tests have historically been the regulatory standard, their correlation with human response varies. Human Repeat Insult Patch Tests (HRIPT) and Human Maximization Tests (HMT) databases containing 2,277 tests across 1,366 substances provide crucial human reference data [43]. These databases help account for interspecies variability and support the validation of non-animal testing approaches. Recent developments include validated positive reference materials like polyurethane sheets containing 1% 2,4-dinitrochlorobenzene (DNCB-PU), which elicit consistent positive responses in both in vivo and in vitro sensitization tests for medical devices [41].
Irritation Testing: Methodologies and Protocols
In Vivo Irritation Assessment Protocols
Irritation testing evaluates the potential of a material or device to cause localized, reversible inflammatory responses at the site of contact. The Primary Skin Irritation Test in rabbits is a standard in vivo method where material extracts or the material itself are applied to shaved rabbit skin under semi-occlusive patches for 4-24 hours. Reactions are scored at various time points for erythema, edema, and other tissue damage using standardized grading scales [40]. The Intracutaneous Reactivity Test provides a more sensitive assessment by injecting material extracts into the rabbit dermis and evaluating the response for evidence of irritation, including hemorrhage, necrosis, or leukocyte infiltration. These in vivo models capture the complex vascular and inflammatory responses that are difficult to fully replicate in vitro, including neurogenic components that contribute to sensations like stinging and burning [40] [42].
Advancements in Alternative Models
Significant progress has been made in developing human cell-based alternatives to animal irritation testing. Reconstructed human epidermis (RhE) models and full-thickness human skin equivalents (HSE) now offer physiologically relevant platforms for irritation assessment [42]. These systems feature stratified squamous epithelium with a functional stratum corneum that mimics skin barrier function. Gene expression profiling has identified specific biomarkers (IL-6, PTGS2, ATF3, TRPV3, MAP3K8, HMGB2, and MMP-3) that can distinguish irritants from non-irritants in these models [42]. While these in vitro approaches cannot fully capture the systemic and sensory components of irritation, they provide valuable screening tools and are increasingly accepted for certain regulatory applications, particularly following European Union directives prohibiting animal testing for cosmetics [42].
Implantation Testing: Methodologies and Protocols
In Vivo Implantation Models and Assessment Criteria
Implantation testing represents the most comprehensive in vivo evaluation for biomaterials intended for surgical applications. These studies assess local tissue effects following device placement in appropriate anatomical sites. The rat subcutaneous or muscle implantation model is commonly used for initial screening, where material samples are surgically implanted and explanted at intervals (e.g., 1, 4, 12, 26 weeks) for histological analysis [7]. More specialized models include bone implantation for orthopedic materials [44] and abdominal wall defects for mesh materials [7]. Histopathological evaluation scores multiple parameters including inflammation severity, fibrosis, necrosis, fatty infiltration, and material degradation. For bone implants, additional endpoints include osteointegration, new bone formation, and osteoclast activity [44].
Table 3: The Scientist's Toolkit: Essential Reagents and Models for In Vivo Evaluations
| Tool Category | Specific Examples | Function in Research | Application Context |
|---|---|---|---|
| Animal Models | Female Sprague-Dawley rats (250-300g) [7], Rodent femoral defect model [44] | Provides intact physiological system for evaluating host response | Implantation studies for soft tissue and bone materials [44] [7] |
| Reference Materials | DNCB-PU sheets [41] | Positive control for sensitization testing | Validation of testing protocols for medical devices [41] |
| Analytical Methods | Histological scoring (0-3 scale) [7], Immunofluorescence for macrophage markers [7] | Quantifies tissue response and cellular infiltration | Evaluation of foreign body reaction and remodeling [7] |
| Cell Culture Systems | Human bone marrow-derived mesenchymal stem cells (hBM-MSCs) [44] | In vitro cytocompatibility assessment prior to in vivo testing | Preliminary biocompatibility screening [44] |
| Imaging Modalities | In vivo and ex vivo X-ray imaging [44] | Monitors gas formation, implant position, and bone healing | Tracking degradation of magnesium implants [44] |
Correlating In Vitro and In Vivo Implantation Outcomes
Research has demonstrated promising approaches for predicting in vivo implantation outcomes using in vitro models. A key advancement involves combining in vitro macrophage-biomaterial interaction assays with in silico analysis methods like principal component analysis (PCA) and dynamic network analysis (DyNA) [7]. These techniques can distinguish materials that will provoke a foreign body reaction (characterized by dominant M1 macrophage response) from those that support constructive remodeling (associated with M2 macrophage polarization) [7]. For example, in vitro analysis of macrophage responses to extracellular matrix (ECM) materials predicted favorable in vivo remodeling outcomes, while responses to synthetic materials like polypropylene and ePTFE predicted foreign body reactions [7]. This approach enables more efficient screening of biomaterial candidates before proceeding to costly in vivo studies.
Integrated Testing Strategies and Future Directions
The future of biomaterial safety testing lies in integrated approaches that combine the strengths of both in vitro and in vivo methodologies. The most effective strategy begins with in vitro screening to eliminate problematic materials early in development, followed by targeted in vivo testing in the most appropriate models [45] [7]. This approach respects the 3Rs principles (Replacement, Reduction, and Refinement) while ensuring comprehensive safety assessment. Technological advances in organ-on-a-chip systems, microphysiological platforms, and sophisticated in silico modeling are increasingly bridging the gap between simple cell cultures and complex living organisms [45] [4]. For implantation studies, the combination of in vitro macrophage response profiling with computational analysis has shown particular promise for predicting in vivo outcomes, potentially reducing the number of animals required for biomaterial development [7]. As these technologies mature, the field is moving toward a more predictive, human biology-based framework for safety assessment while recognizing that in vivo studies remain essential for validating the complex tissue integration and long-term performance of critical medical devices.
The translation of biomaterials from laboratory research to clinical application presents a significant challenge in medical science. At the heart of this challenge lies a critical dilemma: while in vitro (test tube or petri dish) studies provide initial mechanistic insights, they frequently fail to predict complex biological responses that occur in vivo (within a living organism) [25]. This discrepancy was starkly demonstrated in a multicenter analysis of bone regeneration biomaterials, which revealed a "surprisingly poor correlation" between in vitro and in vivo outcomes, with only 58% covariance between experimental results [34]. This translation gap underscores the indispensable role of carefully selected animal models in preclinical biomaterial testing, serving as a crucial bridge between simplified cell cultures and human clinical trials.
The selection of an appropriate animal model represents one of the most consequential decisions in translational biomaterials research [46]. As noted by Bernard Rollin, "The most brilliant design, the most elegant procedures, the purest reagents, along with investigator talent, public money, and animal life are all wasted if the choice of animal is incorrect" [46]. This guide provides a comprehensive comparison of animal models from rodents to non-human primates, offering researchers evidence-based frameworks for model selection aligned with both scientific objectives and ethical considerations.
In Vitro vs. In Vivo Paradigms: Fundamental Distinctions
Defining the Research Approaches
In biomedical research, investigations occur along a spectrum of biological complexity:
In Vitro (Latin for "in glass"): Experiments conducted with biological components (cells, tissues, or biomolecules) in controlled laboratory environments outside living organisms [38] [25]. These include cell culture studies, organ-on-a-chip systems, and biochemical assays performed in petri dishes, test tubes, or multi-well plates.
In Vivo (Latin for "within the living"): Experiments conducted within whole, living organisms [38] [25]. These encompass the entire range of animal studies from rodents to non-human primates, as well as human clinical trials.
Comparative Advantages and Limitations
Table 1: Fundamental Comparisons Between In Vitro and In Vivo Approaches
| Aspect | In Vitro Methods | In Vivo Methods |
|---|---|---|
| Environment | Controlled, simplified artificial systems [38] | Complex, biologically intact living organisms [38] |
| Biological Relevance | Limited to specific cellular interactions [11] | Whole-system responses including immune and endocrine systems [11] |
| Cost & Resources | Relatively low cost, minimal equipment [25] [4] | Very expensive (animal care, monitoring, equipment) [25] [4] |
| Time Efficiency | Rapid results (hours to days) [25] | Lengthy studies (weeks to months) [25] |
| Ethical Considerations | Fewer ethical concerns [11] | Significant ethical considerations, strict oversight required [11] |
| Regulatory Requirements | Less stringent regulations [25] | Strict protocols, IACUC approval required [31] [25] |
| Predictive Value for Human Response | Limited, may not reflect whole-organism physiology [34] [4] | Higher, though species differences persist [25] [4] |
The integration of both approaches creates a powerful iterative research pipeline. The conventional workflow begins with in vitro screening to identify promising candidates, followed by in vivo validation to assess complex physiological responses [38] [4]. As demonstrated in neurogenesis research, in vitro models enable precise manipulation of specific cellular processes, while in vivo models reveal how these processes function within intact biological systems [47].
Rodent Models: Workhorses of Preliminary Biomaterial Testing
Characteristics and Applications
Rodents, particularly mice and rats, represent the most extensively utilized animal models in preliminary biomaterial research. Their popularity stems from practical advantages including small size, ease of handling, rapid reproduction cycles, and well-characterized genetics [4]. Mice share approximately 95% of their genes with humans, making them particularly valuable for genetic studies and preliminary efficacy testing [4].
In bone regeneration research, rodent models provide essential first-line in vivo data on biomaterial integration, biodegradation, and early host inflammatory responses [34]. The rat skeletal muscle injury model has been effectively used to evaluate the host response to various biomaterials, assessing macrophage polarization and tissue remodeling outcomes [7].
Limitations in Translational Predictability
Despite their utility, rodent models present significant limitations in predicting human responses to biomaterials. Key physiological differences include:
- Metabolic variations: Differential drug metabolism and clearance rates compared to humans [25]
- Size disparities: Substantial differences in body mass and organ scale affecting dosage calculations [46]
- Cellular composition: Variations in neural stem cell populations and cortical development compared to primates [47]
These species-specific differences frequently manifest in failed translations, where biomaterials demonstrating efficacy in rodent models prove ineffective or unsafe in human trials [46].
Non-Human Primate Models: Bridging the Translational Gap
Physiological Similarities to Humans
Non-human primates (NHPs), including monkeys and apes, represent the most physiologically proximate models to humans, sharing significant anatomical, genetic, and functional similarities [47]. These similarities are particularly pronounced in specialized biological systems:
- Neurogenesis patterns: Primates exhibit expanded cortical regions with slower neuroepithelial to radial glia transitions and presence of a distinctive outer subventricular zone (OSVZ) enriched with outer radial glia cells [47]
- Neural development: Unlike rodents, primates possess abundant outer radial glia and basal intermediate progenitors during development, both displaying high proliferative capacity [47]
- Subplate zone expansion: Experience secondary expansion of the subplate zone during cerebral development, similar to humans [47]
- Immune system function: Greater similarity in immune response patterns compared to rodents [47]
Specific Research Applications
NHP models provide critical insights for biomaterial applications requiring species-specific biological responses. In neurological biomaterial research, NHPs enable evaluation of interventions in gyrencephalic brains (with cortical folds) rather than the lissencephalic (smooth) brains of rodents [47]. The expanded subventricular zone and diverse neural progenitor populations in NHPs make them indispensable for testing biomaterials designed for neural regeneration applications [47].
Direct Comparison: Rodents versus Non-Human Primates
Table 2: Systematic Comparison of Rodent and Non-Human Primate Models for Biomaterial Research
| Parameter | Rodent Models | Non-Human Primate Models |
|---|---|---|
| Phylogenetic Proximity to Humans | Distant (∼95% gene similarity in mice) [4] | Close (significant anatomical/functional similarities) [47] |
| Cortical Neurogenesis | Limited outer subventricular zone, scarce outer radial glia [47] | Expanded outer subventricular zone, abundant outer radial glia [47] |
| Brain Architecture | Lissencephalic (smooth brain) [47] | Gyrencephalic (folded brain) [47] |
| Research Costs | Relatively low [4] | Very high (housing, care, procedures) [25] |
| Regulatory Oversight | Standard animal research protocols | Enhanced oversight, stricter justification requirements [48] |
| Public Perception | Generally accepted for research | Heightened ethical concerns [25] |
| Experimental Timeline | Short (rapid maturation and reproduction) [4] | Long (extended maturation, single offspring) [47] |
| Typical Applications | Preliminary efficacy, mechanism of action, initial safety [4] | Final preclinical validation, complex system interactions [47] |
Methodological Framework for Animal Model Selection
Experimental Design Considerations
Selecting the appropriate animal model requires systematic consideration of multiple scientific and practical factors:
- Research question alignment: Does the model's biology appropriately address the specific research question? [46]
- Species-specific physiological relevance: Are the relevant physiological processes sufficiently similar to humans? [46]
- Feasibility and resources: Are the necessary expertise, facilities, and funding available for proper model implementation? [46]
- Ethical justification: Can the use of a higher species be sufficiently justified, following the 3R principles (Replacement, Reduction, Refinement)? [48]
- Statistical power: Has the appropriate number of animals been calculated to reach statistical significance while minimizing subjects? [46]
Integrating the 3R+DQ Framework
Modern biomaterials research emphasizes the 3R+DQ framework—Replacement, Reduction, Refinement, plus Design and Quality [48]. This approach mandates:
- Replacement: Using in vitro or computational methods whenever possible to replace animal use [48]
- Reduction: Implementing statistical methods to minimize animal numbers while maintaining scientific validity [46]
- Refinement: Enhancing experimental techniques to minimize pain and distress [48]
- Design and Quality: Ensuring rigorous study design and execution under quality management systems [48]
Figure 1: Animal Model Selection Workflow. This decision pathway illustrates the sequential integration of experimental models in biomaterial development.
Experimental Protocols and Methodologies
Standardized Biomaterial Implantation Protocol
The following methodology, adapted from published studies, outlines a standardized approach for evaluating biomaterials in rodent models [7]:
- Animal Preparation: Female Sprague-Dawley rats (250-300 g) are acclimated for 7 days with ad libitum access to food and water [7]
- Surgical Procedure:
- Anesthesia induction and maintenance using appropriate agents (e.g., isoflurane)
- Ventral abdominal incision approximately 2 cm lateral to midline
- Creation of paramedian 1×1 cm² partial-thickness abdominal wall defect
- Excision of external and internal oblique muscles while preserving transversalis fascia
- Biomaterial Implantation:
- Size-matched device coupons (1×1 cm²) inlaid within defect
- Fixation to adjacent muscle at four corners with non-degradable 4-0 polypropylene interrupted sutures
- Skin closure with degradable 4-0 Vicryl sutures
- Post-operative Monitoring:
- Analgesia administration for 48-72 hours
- Daily observation for signs of distress or complications
- Endpoint Analysis:
- Euthanasia at predetermined timepoints (e.g., 14, 35 days)
- Explanation of implants with surrounding tissue
- Fixation in neutral buffered formalin
- Processing for histological analysis (dehydration, paraffin embedding, 5-μm sectioning)
Host Response Evaluation Methods
Comprehensive assessment of host-biomaterial integration involves multiple analytical techniques:
- Histological Evaluation: Hematoxylin and eosin staining for general tissue architecture and cellular infiltration [7]
- Histomorphological Scoring: Semiquantitative analysis (0-3 scale) of:
- Foreign body giant cell formation
- Biomaterial degradation
- Connective tissue organization
- Encapsulation
- Muscle ingrowth
- Immunofluorescence Characterization:
- Macrophage polarization markers (CCR7 for M1 phenotype; CD206 for M2 phenotype) [7]
- Secondary antibodies with fluorescent conjugates for visualization
- Confocal microscopy for image acquisition
- Quantitative analysis of cell populations
Essential Research Reagents and Materials
Table 3: Essential Research Reagents for Biomaterial Animal Studies
| Reagent/Material | Function/Application | Specific Examples |
|---|---|---|
| Synthetic Biomaterials | Mesh devices for soft tissue reconstruction | Polypropylene, polyglycolide, polyethylene, ePTFE [7] |
| ECM-Derived Materials | Biologically active scaffolds for constructive remodeling | Urinary bladder matrix, dermal ECM, small intestinal submucosa [7] |
| Suture Materials | Surgical implantation and wound closure | Polypropylene (non-degradable), Vicryl (degradable) [7] |
| Histological Stains | Tissue structure visualization | Hematoxylin and eosin [7] |
| Immunofluorescence Reagents | Cell phenotype identification | Primary antibodies (CCR7, CD206), fluorescent secondary antibodies [7] |
| Decellularization Agents | ECM preparation from source tissues | Trypsin, ethanol, Triton X-100, peracetic acid [7] |
| Crosslinking Compounds | Biomaterial modification | Ethyldimethylaminopropyl carbodiimide (EDAC) [7] |
Emerging Approaches and Future Directions
Advanced Predictive Methodologies
Innovative approaches are emerging to enhance the predictive value of preclinical testing:
- In vitro-in vivo correlation (IVIVC): Combining in vitro macrophage assays with in silico analysis (principal component analysis and dynamic network analysis) to predict in vivo host responses [7]
- Organ-on-a-chip technologies: Microfluidic devices that simulate human organ physiology for more relevant in vitro screening [4]
- Humanized animal models: Genetically modified rodents expressing human genes or hosting human tissues to better mimic human responses
Refined Model Selection Strategies
Future biomaterials research will likely embrace more nuanced model selection strategies:
- Disease-specific model matching: Aligning particular disease pathophysiology with appropriate animal models that faithfully recapitulate key aspects [46]
- Staged model deployment: Implementing sequential use of models based on increasing complexity and information needs [47]
- Enhanced reporting standards: Improving documentation of model characteristics to enable better cross-study comparisons and meta-analyses
Figure 2: Evolution of Animal Model Selection Strategies in Biomaterials Research
The selection of appropriate animal models represents a critical determinant of success in biomaterials research. Rodent models provide accessible, cost-effective platforms for preliminary efficacy and safety assessment, while non-human primate models offer unparalleled physiological relevance for final preclinical validation of complex biological responses. The documented poor correlation between in vitro and in vivo testing [34] underscores the continued necessity of whole-animal models, despite advances in alternative technologies.
Researchers must approach model selection as a strategic decision, carefully balancing scientific objectives, ethical responsibilities, and practical constraints. By implementing the 3R+DQ framework [48] and selecting models based on specific research questions rather than convenience, the scientific community can enhance the translational value of preclinical biomaterial testing while upholding the highest standards of animal welfare. Through thoughtful model selection and rigorous experimental design, researchers can bridge the translational gap and accelerate the development of innovative biomaterials for clinical application.
In the fields of orthopedics and cardiovascular medicine, the performance of implantable materials directly determines clinical outcomes and patient quality of life. Biomaterials must demonstrate not only excellent mechanical properties but also biocompatibility, durability, and the ability to integrate with host tissues without provoking adverse reactions. The evaluation of these materials spans from controlled in vitro laboratory studies to clinical registries that track real-world performance in diverse patient populations. This guide objectively compares the performance of various biomaterials used in these specialties, with a specific focus on the critical relationship between standardized in vitro testing results and long-term in vivo outcomes. Understanding this relationship is essential for researchers, regulatory bodies, and clinicians who must predict clinical performance from pre-market data and identify potential safety issues before they affect large patient cohorts.
Performance Benchmarking of Orthopedic Biomaterials
Material Classes and Key Characteristics
Orthopedic implants require a combination of high strength, wear resistance, and excellent biocompatibility. The table below compares the primary material classes used in orthopedics, such as joint replacements and fracture fixation devices [49].
Table 1: Comparison of Major Orthopedic Biomaterial Classes
| Material Class | Specific Examples | Key Advantages | Key Limitations & Biocompatibility Concerns |
|---|---|---|---|
| Stainless Steel | 316L (Low carbon) | High ductility, well-established processing, low cost | Susceptibility to crevice corrosion; release of nickel and chromium ions can cause sensitivity (affects ~20% of population) and potential toxicity [49]. |
| Cobalt-Chromium Alloys | CoCrMo | Excellent wear resistance, high strength | Potential for metal ion release; concerns regarding biological response to cobalt and chromium [49]. |
| Titanium Alloys | Ti6Al4V | Superior corrosion resistance, excellent biocompatibility, elastic modulus closer to bone | Poor wear resistance; requires surface modification for articulating surfaces [50] [49]. |
| Polyethylene | Ultra-High-Molecular-Weight Polyethylene (UHMWPE) | High toughness, low friction | Wear debris can lead to osteolysis (bone resorption) and implant loosening [49]. |
| Ceramics | Alumina, Zirconia | Exceptional wear resistance, inertness, high compressive strength | High stiffness, brittleness leading to risk of fracture [49]. |
| Biodegradable Metals | Magnesium alloys (e.g., Mg-Sr) | Biodegradable, avoids second surgery, biocompatible | Degradation rate must match bone healing process; rapid corrosion can hinder healing [50]. |
Case Study: The "Metal-on-Metal" (MoM) Hip Implant Failure
A pivotal case in orthopedic benchmarking was the failure of certain MoM hip implants. Initially marketed as a more durable alternative, mid-term follow-up data from national joint replacement registries identified far higher revision rates compared to other bearing surfaces [51]. For instance, the Australian Orthopaedic Association National Joint Replacement Registry flagged these devices as performance outliers a full three years before their market withdrawal [51]. This case underscores the irreplaceable role of comprehensive post-market surveillance in identifying safety issues that may not be apparent in limited pre-clinical trials.
Performance Benchmarking of Cardiovascular Biomaterials
Material Classes and Applications
Cardiovascular materials are used in devices like stents, valves, and vascular grafts, where they must function in a dynamic mechanical and biological environment. The following table summarizes key materials and their performance considerations [50] [52].
Table 2: Comparison of Major Cardiovascular Biomaterial Classes and Applications
| Material Class | Common Applications | Performance Advantages | Performance Limitations & Clinical Challenges |
|---|---|---|---|
| Metallic Alloys | Stents (Coronary, peripheral), Valve frames, Guidewires | Titanium (Ti) & Cobalt-Chromium (Co-Cr) offer high strength and radial force. | Overstable permanent implants (e.g., Ti) may require secondary surgery. Thrombogenicity (clot risk) necessitates drug coatings or anticoagulation [50] [52]. |
| Biodegradable Metals | Stents (e.g., Mg-based, Iron-based) | Eliminates permanent implant, potentially reduces long-term complications. | Degradation rate control is critical. Mg may degrade too quickly, while Fe may degrade too slowly. Degradation products must be managed [50]. |
| Biological Tissues | Bioprosthetic Heart Valves (from bovine pericardium, porcine valves) | Superior hemocompatibility, avoids lifelong anticoagulation. | Prone to calcification and structural deterioration, especially in younger patients. Limited long-term durability [52]. |
| Polymer Scaffolds | Tissue-Engineered Valves, Vascular Grafts (e.g., PCL, PLA) | Allows for in situ tissue engineering; biodegradable, off-the-shelf availability. | Requires a precise balance between scaffold degradation and neo-tissue formation. Long-term durability under physiological forces is under investigation [52]. |
| Decellularized Scaffolds | Patches, Valves (e.g., SynerGraft, CorMatrix, CardioCel) | Provides a natural ECM structure for host cell repopulation. | Can provoke inflammatory response, leading to calcification or early failure (e.g., early decellularized porcine valves). Outcomes can be unpredictable [52]. |
Case Study: Coronary Stent Thrombosis
Cardiovascular device registries have proven invaluable for monitoring the safety of high-risk devices like coronary stents. These registries have documented increased rates of rare but serious adverse events, such as stent thrombosis, with specific stent platforms [51]. By capturing data from an unselected, population-based cohort, registries provide the statistical power necessary to detect these low-frequency safety signals, which are often missed in pre-market clinical trials with smaller sample sizes and limited follow-up duration.
In Vitro vs. In Vivo Testing: A Comparative Analysis
The journey of a biomaterial from the lab to the clinic involves a multi-stage validation process. The following diagram illustrates the typical workflow and the key questions asked at each stage, highlighting the transition from controlled in vitro experiments to complex in vivo environments and post-market surveillance.
Diagram 1: Biomaterial Testing Pathway
In Vitro Testing Protocols and Standards
In vitro testing provides the first line of evaluation for new biomaterials, assessing fundamental properties like cytotoxicity and mechanical strength under controlled conditions.
Cytotoxicity Testing (ISO 10993-5)
Objective: To evaluate the potential of a material or its extracts to cause cell death or inhibit cell growth. This is a mandatory test for regulatory approval of new medical devices [30].
Detailed Protocol (MTT Assay for Mg-1%Sn-2%HA Composite):
- Sample Preparation: The test material (e.g., Mg-1%Sn-2%HA composite) is sterilized. Using an elution method, an extract is prepared by immersing the material in a cell culture medium (e.g., Dulbecco’s Modified Eagle Medium supplemented with fetal bovine serum) for a specified period (e.g., 24 hours) at 37°C [30].
- Cell Culture: L-929 mouse fibroblast cells (a standard cell line) are cultured and seeded into multi-well plates and allowed to attach [30].
- Exposure: The culture medium is replaced with the prepared extract. Cells are incubated with the extract for a defined duration (e.g., 7 days) at 37°C with 5% CO₂ [30].
- Viability Assessment: After incubation, the MTT reagent is added. Mitochondrial dehydrogenases in viable cells convert the yellow MTT into purple formazan crystals. The crystals are dissolved using an organic solvent (e.g., isopropanol), and the intensity of the purple color, which is proportional to the number of viable cells, is measured using a spectrophotometer at 492 nm [30].
- Data Analysis: Cell viability is calculated as a percentage compared to untreated control cells. According to ISO 10993-5, a cell viability greater than 70-80% is typically required to classify a material as non-cytotoxic. In the cited study, the Mg-1%Sn-2%HA composite showed 71.51% viability with the undiluted extract, confirming its non-cytotoxic status [30].
In Vivo and Post-Market Performance Benchmarking
While in vitro tests are essential for screening, in vivo and post-market studies reveal how materials perform in the complex reality of the human body.
The Role of Medical Device Registries in Post-Market Surveillance
Objective: To continuously collect real-world data on the safety and performance of medical devices in an unselected patient population after they have received regulatory approval and CE marking or FDA clearance [51].
Detailed Protocol (European Cardiovascular and Orthopaedic Registries):
- Data Collection: Regional, national, or multi-country registries systematically collect relevant data on device implantation. This includes patient demographics, device identifier (e.g., Unique Device Identification), procedural details, and outcomes [51].
- Quality Metrics: The utility of a registry depends on key quality principles:
- Coverage: The extent of participation in data collection across the target population.
- Completeness: The proportion of data consistently captured for all procedures.
- Accuracy: The degree to which recorded data reflects the actual healthcare event.
- Standardization: The use of uniform outcome definitions and follow-up durations [51].
- Data Analysis and Outlier Detection: Registry data is analyzed to benchmark device performance. This involves calculating rates of adverse events (e.g., revision surgery, stent thrombosis) and statistically identifying devices that are "outliers" with significantly worse performance than the average [51].
- Reporting: Findings are disseminated through annual reports and peer-reviewed publications to inform clinicians, regulators, and manufacturers.
Benchmarking Data from European Registries: A systematic review of European cardiovascular and orthopaedic registries revealed significant heterogeneity in their structure and reporting [51]. Key findings include:
- Data Completeness: A median of 60% of key quality items were reported by orthopaedic registries, compared to only 33% by cardiovascular registries [51].
- Outlier Reporting: None of the cardiovascular registries reported on outlier performance of devices, while 19% (5 out of 26) of orthopaedic registries did so, albeit with different outlier definitions [51].
- Follow-up and Outcomes: There was large heterogeneity in reporting on outcomes, definitions of outcomes, and follow-up durations across all registries [51].
Table 3: In Vitro vs. In Vivo/Registry Testing: A Comparative Summary
| Aspect | In Vitro Testing | In Vivo / Registry Data |
|---|---|---|
| Environment | Controlled, simplified | Complex, dynamic biological system |
| Key Metrics | Cytotoxicity, mechanical properties, degradation in simulated fluid | Long-term survival/revision rates, device-specific complications (e.g., thrombosis), patient-reported outcomes |
| Regulatory Role | Pre-market screening for biocompatibility and safety | Post-market surveillance for long-term safety and performance |
| Strengths | Rapid, low-cost, high-throughput, ethically simpler | Provides real-world evidence, detects rare/long-term adverse events |
| Limitations | May not predict full host response (immune system, healing) | Expensive, requires large patient numbers, data quality can be variable |
| Case Study Link | Mg-1%Sn-2%HA composite shows high cell viability [30] | Metal-on-Metal hip implants identified as outliers by registries [51] |
The Scientist's Toolkit: Essential Research Reagents and Materials
The following table details key reagents, materials, and tools used in the experiments and fields discussed in this guide.
Table 4: Essential Research Reagents and Materials for Biomaterial Testing
| Item | Function / Application | Relevant Context |
|---|---|---|
| L-929 Mouse Fibroblast Cells | A standard cell line used for in vitro cytotoxicity testing according to ISO 10993-5. | Cytotoxicity testing of the Mg-1%Sn-2%HA composite [30]. |
| MTT Assay Kit | A colorimetric assay to measure cell viability and proliferation based on mitochondrial activity. | Quantifying the cytotoxicity of material extracts [30]. |
| ISO 10993-5 Standard | The international standard that specifies test methods to assess the in vitro cytotoxicity of medical devices. | Protocol foundation for all cytotoxicity testing of new medical devices [30]. |
| Polylactic Acid (PLA) | A biodegradable thermoplastic polymer used in 3D printing of scaffolds for tissue engineering. | Neural tissue engineering, cardiovascular scaffolds [52] [53]. |
| Polycaprolactone (PCL) | A biodegradable polyester with good mechanical elasticity, used in fused deposition modeling (FDM) 3D printing. | Neural guide conduits, cardiovascular implants [52] [53]. |
| Gelatin Methacrylate (GelMA) | A photocrosslinkable hydrogel derived from gelatin; provides a biomimetic environment for cell growth. | Used in 3D bioprinting for creating cell-laden scaffolds in neural and other tissue engineering applications [53]. |
| Medical Device Registry | An organized system for continuous collection of data on medical devices in a defined population. | Post-market surveillance of cardiovascular and orthopaedic implants (e.g., coronary stents, hip prostheses) [51]. |
The benchmarking of orthopedic and cardiovascular materials reveals a critical and often complex relationship between in vitro predictions and in vivo performance. While standardized in vitro tests like cytotoxicity assays are indispensable for initial screening and regulatory approval, they cannot fully replicate the long-term, dynamic biological environment. As demonstrated by the Mg-1%Sn-2%HA composite, a material can perform excellently in the lab, but its real-world success depends on a harmonious balance between degradation and tissue regeneration in the body [30] [50]. Conversely, the failures of MoM hip implants and certain decellularized cardiovascular patches highlight the irreplaceable role of high-quality, standardized device registries in identifying safety issues that emerge only after widespread clinical use [51] [52]. The future of biomaterial development lies in leveraging advanced tools like artificial intelligence for property prediction and 3D bioprinting for creating more physiologically relevant in vitro models [54]. However, closing the translation gap will require a continued commitment to robust post-market clinical follow-up and international harmonization of registry data to ensure that the performance benchmarks set in the laboratory are consistently met in the clinic.
In the field of biomaterial development, interpreting test endpoints from cellular assays to tissue-level analysis represents a critical juncture in translating preclinical findings to clinical applications. The evaluation chain, spanning from isolated cell viability studies to comprehensive histopathological assessment, provides the essential data for determining biomaterial safety and efficacy. However, this pathway is marked by significant interpretive challenges, as results from controlled in vitro environments frequently demonstrate poor predictive value for outcomes in complex living systems [34]. This guide objectively compares these fundamental testing approaches, examining their respective methodologies, performance characteristics, and the critical factors influencing correlation between their endpoints. Framed within broader research on in vitro versus in vivo testing comparisons, this analysis provides drug development professionals and researchers with a structured framework for selecting appropriate testing strategies and interpreting their results within a cohesive biological context.
Quantitative Correlation Between In Vitro and In Vivo Outcomes
A cornerstone challenge in biomaterial testing lies in the discordance between results obtained in controlled laboratory environments and those observed in whole living organisms. A comprehensive multicentre analysis provides compelling quantitative evidence of this disconnect, revealing surprisingly poor correlation between established in vitro assays and in vivo outcomes for bone regeneration biomaterials [34].
Table 1: Correlation Analysis Between In Vitro and In Vivo Testing Outcomes
| Assessment Type | Number of Studies Analyzed | Number of Biomaterials Tested | Correlation Finding | Statistical Significance |
|---|---|---|---|---|
| Overall Correlation | 36 in vivo + 47 in vitro | 93 different biomaterials | No significant overall correlation | Not statistically significant |
| Mean Score Trend | Data from all provided studies | 93 different biomaterials | 58% covariance trend | Trend observed, not significant |
This extensive review, which incorporated data from 36 in vivo studies and 47 in vitro assays across 8 European universities, found no statistically significant overall correlation between in vitro and in vivo assessments [34]. While a trend of covariance (58%) was observed in mean scores, the findings highlight the substantial limitations of current in vitro methodologies in predicting in vivo performance. The authors concluded that this "surprisingly poor correlation" indicates "a clear need for further development of relevant in vitro assays" and "validated pre-clinical pipelines" to improve predictive accuracy [34].
Experimental Protocols for Key Testing Methodologies
In Vitro Cytotoxicity Testing (ISO 10993-5)
Cytotoxicity testing represents a fundamental component of the "Big Three" biocompatibility tests required for nearly all medical devices, alongside irritation and sensitization assessment [21]. The standardized protocol evaluates whether device materials or their extracts cause harm to living cells.
Detailed Methodology:
- Extract Preparation: Biomaterials are immersed in extraction solvents (e.g., physiological saline, vegetable oil, cell culture medium) under specified conditions [21].
- Cell Culture Exposure: Cultured mammalian cell lines (e.g., Balb 3T3, L929, or Vero fibroblasts) are exposed to device extracts for approximately 24 hours [21].
- Endpoint Assessment: Multiple endpoints are evaluated after the exposure period:
- Cell Viability: Measured using quantitative assays including MTT, XTT, or neutral red uptake [21].
- Morphological Changes: Documented alterations in cell shape or structure.
- Cell Detachment: Degree of cell detachment from culture substrate.
- Cell Lysis: Presence of cellular debris or membrane damage.
- Acceptance Criteria: While ISO 10993-5 does not define strict acceptance criteria, ≥70% cell survival (viability) is generally considered a positive indicator, particularly when testing neat extract [21].
In Vivo Histopathological Analysis
Histopathological analysis provides critical information about tissue responses to implanted biomaterials in a living system, offering insights not attainable through in vitro methods. This protocol typically involves implantation in animal models followed by microscopic examination of tissue responses.
Detailed Methodology:
- Study Design: Biomaterials are implanted into appropriate animal models (e.g., rodents) for predetermined durations based on the intended application [55].
- Tissue Harvesting: After sacrifice at designated time points, implanted tissues and surrounding areas are harvested.
- Tissue Processing: Samples undergo fixation, dehydration, clearing, infiltration, and embedding in paraffin blocks.
- Sectioning and Staining: Thin sections (typically 5-10μm) are cut and mounted on slides, then stained with Hematoxylin and Eosin (H&E) to visualize tissue morphology and cellular structure [56].
- Microscopic Evaluation: Trained pathologists examine stained sections for key tissue response parameters:
- Acute Inflammation: Characterized by presence of polymorphonuclear leukocytes (neutrophils).
- Chronic Inflammation: Identified by monocytes and lymphocytes.
- Foreign Body Reaction: Macrophages and foreign body giant cells at tissue-implant interface.
- Granulation Tissue: Fibroblasts and new blood vessels indicating healing response.
- Fibrous Encapsulation: Fibroblasts and collagen deposition forming a capsule around the implant [55].
- Scoring System: Responses are typically scored using semi-quantitative scales based on cellular density, tissue integrity, and specific cell type presence.
Advanced Techniques: Automated Cell Classification
Emerging technologies are addressing limitations in traditional histopathological analysis. Automated cell annotation using multiplexed immunofluorescence (mIF) with H&E staining represents a significant advancement, overcoming inter-observer variability in manual annotations where pathologist agreement for identifying macrophages can be as low as 50% [57].
Detailed Methodology:
- Serial Staining: Tissue sections undergo mIF staining for cell lineage protein markers (e.g., pan-CK, CD3, CD20, CD66b, CD68) followed by H&E staining of the same section [57].
- Image Co-registration: Precise alignment of mIF and H&E images at single-cell level, achieving average cell-cell distance of 3.1 microns (below average nucleus size of 7.6 microns) [57].
- Cell Type Definition: Unsupervised clustering (Leiden algorithm) of protein marker intensity values defines cell types objectively, without arbitrary manual gating cutoffs.
- Model Training: Deep learning models trained on mIF-labeled H&E images achieve 86%-89% accuracy in classifying four major cell types (tumor cells, lymphocytes, neutrophils, macrophages) [57].
- Spatial Biomarker Discovery: Single-cell analysis enables identification of spatial interactions between immune cells linked to clinical outcomes and therapeutic responses.
Comparative Analysis of Testing Approaches
Performance Characteristics Across Testing Modalities
Table 2: Comparative Analysis of Testing Method Performance
| Characteristic | In Vitro Cytotoxicity | Traditional Histopathology | AI-Enhanced Histopathology |
|---|---|---|---|
| Testing Environment | Controlled laboratory setting [58] | Living organism [58] | Living organism with computational analysis [57] |
| Primary Endpoints | Cell viability, morphological changes [21] | Tissue response continuum, inflammation, fibrosis [55] | Single-cell classification, spatial relationships [57] |
| Temporal Resolution | Short-term (typically 24-72 hours) [21] | Multiple time points (acute to chronic) [55] | Multiple time points with single-cell resolution [57] |
| Throughput | High-throughput, scalable [59] | Low-throughput, time-consuming | High-throughput once model trained [57] |
| Predictive Value for Clinical Outcomes | Limited correlation (58% covariance) [34] | Moderate to high, but subjective | Potentially higher, objective quantification [57] |
| Key Limitations | Cannot replicate whole-organism complexity [34] [58] | Inter-observer variability, semi-quantitative [56] | Requires complex validation, specialized expertise [57] |
Concordance and Discordance in Endpoint Interpretation
Significant discordance exists between different assessment methods, highlighting the importance of complementary testing approaches:
- Visual vs. Molecular Assessment: In burn wound evaluation, substantial discordance exists between intraoperative visual assessment and molecular determination of regenerative capacity. Lactate dehydrogenase, Keratin-15, and Ki67 markers revealed significant viability and regenerative capacity in tissue clinically assessed as non-viable, demonstrating how visual assessment can lead to removal of potentially salvageable tissue [56].
- Histological Limitations: H&E staining alone shows no differences in staining between viable and non-viable tissue, limiting its utility as a standalone assessment method for determining tissue viability and regenerative capacity [56].
- Cellular vs. Tissue-Level Responses: In vitro cytotoxicity may fail to predict in vivo implantation responses due to differences in cell types used (often tumor-derived fibroblasts) versus the specific cells encountered in vivo (e.g., macrophages in foreign body response) [55].
Essential Research Reagent Solutions
Table 3: Key Research Reagents for Test Endpoint Analysis
| Reagent Category | Specific Examples | Primary Function | Application Context |
|---|---|---|---|
| Cell Viability Assays | MTT, XTT, Neutral Red Uptake | Quantitative measurement of cell metabolic activity and viability [21] | In vitro cytotoxicity testing per ISO 10993-5 [21] |
| Cell Line Markers | Keratin-15, Ki67 | Identification of epidermal progenitor cells and proliferative capacity [56] | Assessment of regenerative capacity in wound healing models [56] |
| Metabolic Activity Markers | Lactate Dehydrogenase (LDH) | Enzymatic assessment of cellular viability and membrane integrity [56] | Determination of viable tissue in burn wounds and other injury models [56] |
| Immunofluorescence Markers | CD3, CD20, CD66b, CD68, pan-CK | Cell lineage identification for automated classification [57] | Multiplexed immunofluorescence for objective cell typing [57] |
| Histological Stains | Hematoxylin and Eosin (H&E) | Visualization of tissue morphology and cellular structure [56] | Standard histopathological assessment of tissue response [56] |
The interpretation of test endpoints from cell viability to histopathological analysis requires careful consideration of the strengths and limitations inherent to each methodology. While in vitro approaches offer controlled, scalable systems for initial screening, their predictive value for in vivo outcomes remains limited, with multicentre analyses demonstrating only 58% covariance between these testing domains [34]. Conversely, traditional histopathology provides crucial whole-system context but suffers from subjectivity and inter-observer variability. Emerging technologies, particularly AI-enhanced automated cell classification, offer promising avenues for bridging this gap through objective, quantitative analysis of cellular responses within tissue contexts [57]. For comprehensive biomaterial evaluation, researchers should employ integrated testing strategies that leverage the complementary strengths of both in vitro and in vivo approaches while acknowledging the current limitations in correlating their endpoints. This multifaceted approach provides the most robust foundation for predicting clinical performance and advancing biomaterial development.
Overcoming Discrepancies: Strategies to Improve Test Predictivity
In pharmaceutical research and development, the disconnect between experimental results obtained in the laboratory and outcomes observed in living organisms represents one of the most significant challenges. This "correlation crisis" has profound implications, with approximately 90% of drug candidates failing during clinical development despite promising preclinical data [60] [61]. Analysis of clinical trials from 2010-2017 reveals that 40-50% of failures stem from lack of clinical efficacy, while approximately 30% result from unmanageable toxicity [60] [62]. This high failure rate persists despite implementation of rigorous validation processes, highlighting the critical need to understand and address the fundamental discrepancies between in vitro and in vivo systems.
The financial and temporal costs of this translation gap are staggering. Bringing a single drug to market typically requires 10-15 years and over $2 billion, with a substantial portion invested in candidates that ultimately fail in clinical trials [61]. This review examines the biological and methodological factors underlying the correlation crisis, compares current testing methodologies, presents experimental data demonstrating these disparities, and explores emerging strategies to enhance the predictive power of preclinical research.
Fundamental Differences Between In Vitro and In Vivo Systems
Definition and Methodological Comparison
In vitro (Latin for "in glass") refers to experimentation conducted with microorganisms, cells, or biological molecules outside their normal biological context, in controlled environments such as petri dishes or test tubes [2]. In contrast, in vivo (Latin for "within the living") involves research conducted on whole, living organisms, including animal studies and human clinical trials [2] [63]. A third category, in silico studies, comprises biological experiments carried out entirely via computer simulation [63].
Table 1: Core Differences Between Experimental Approaches
| Parameter | In Vitro | In Vivo | In Silico |
|---|---|---|---|
| Complexity | Isolated cells or components | Whole living organisms | Computer simulations |
| Control | High environmental control | Limited control over variables | Complete parameter control |
| Cost & Throughput | Lower cost, higher throughput | High cost, lower throughput | Lowest cost, highest throughput |
| Biological Relevance | Limited, reductionist | High, captures system complexity | Model-dependent |
| Regulatory Status | Early screening | Required for safety/effi cacy | Supplemental support |
Key Biological Factors Contributing to the Correlation Gap
The translation crisis stems from fundamental biological differences that in vitro systems cannot fully recapitulate:
Absence of Systemic Physiology: In vitro models lack the endocrine, neural, and immune system interactions present in whole organisms [64]. They cannot replicate the complex interplay between different organs and tissues that determines drug distribution, metabolism, and overall pharmacological effect [63].
Barrier Penetration Limitations: In vitro systems typically cannot model the complex biological barriers that drugs must cross in vivo, such as the blood-brain barrier, intestinal epithelium, or cellular membranes with their specific transport systems [62].
Protein Binding and Distribution: The distribution of compounds in vivo is significantly influenced by binding to serum proteins and tissue components, which affects drug availability and clearance [62]. In vitro systems often poorly predict these distribution patterns.
Metabolic Complexity: While simple metabolic systems can be modeled in vitro, they cannot replicate the full metabolic capacity of a living organism, particularly the interplay between hepatic metabolism, biliary excretion, and enterohepatic recirculation [65].
Quantitative Evidence of the Disconnect
Immunotoxicity Assessment Disparities
The field of nanomedicine provides compelling examples of correlation challenges. Standard in vivo toxicity tests demonstrate low sensitivity to immunotoxicities, complicated by inter-species variability in immune system structure and function [64]. This has created an urgent need for trustworthy in vitro assays that can better predict human immune responses.
Table 2: In Vitro vs. In Vivo Correlation Challenges in Immunotoxicity Testing
| Immunotoxicity Parameter | In Vitro Challenges | In Vivo Translation Issues |
|---|---|---|
| Hemolysis | Direct measurement possible | Complex clearance mechanisms |
| Complement Activation | Can detect activation | May not predict anaphylaxis severity |
| Thrombogenicity | Platelet activation measurable | Vascular thrombosis complexity |
| Cytokine Induction | Inflammatory response detectable | May not predict cytokine storm |
| Phagocytosis | Uptake measurable | Clearance by MPS affects distribution |
Analysis of drug withdrawal data reveals that 10-20% of drugs withdrawn from clinical use between 1969 and 2005 were removed due to immunotoxicity issues, including anaphylaxis, allergy, hypersensitivity, and immunosuppression [64]. This suggests significant gaps in preclinical immunotoxicity prediction.
Pharmacokinetic Prediction Challenges
In Vitro-In Vivo Extrapolation (IVIVE) approaches that use in vitro metabolism data to predict human drug clearance systematically underestimate actual in vivo results with 3- to 10-fold errors [65]. This consistent underestimation persists despite model optimization efforts, though the refined approaches can achieve under-prediction of only 1.25-fold for hepatocyte assays and 3.5-fold for microsome stability assays [65].
The well-stirred model, one of the simplest and most widely used predictive models for early screening of new chemical entities, demonstrates these systematic prediction errors [65]. Even with advanced optimization, these models struggle to accurately predict human clearance, particularly for compounds affected by transporter proteins or complex distribution patterns.
Methodological Limitations and Experimental Considerations
Critical Factors in Experimental Design
Several methodological factors significantly impact the predictive value of in vitro studies:
Nanoparticle Interference with Assays: In nanomedicine, nanoparticles can interfere with in vitro immunoassays, producing false results [64]. The vast variety of engineered nanomaterials complicates establishing harmonized testing protocols.
Concentration Selection: Choosing appropriate nanoparticle concentrations for in vitro tests to ensure predictive value for in vivo toxicity remains challenging [64]. The selected concentration must balance physiological relevance with detection capabilities.
Control Selection: Identifying appropriate positive and negative controls for nanoformulations is complicated by the assumption that different particle types may behave differently in a given assay [64]. A liposome's toxicity mechanism may differ fundamentally from that of a metal oxide nanoparticle.
Species Specificity: Inter-species variability in the structure and function of the immune system limits the predictive value of animal models for human responses [64]. This is particularly relevant for immunotoxicity assessment.
The Research Reagent Solution Toolkit
Table 3: Essential Research Tools for In Vitro - In Vivo Correlation Studies
| Reagent/Model System | Function | Application Examples |
|---|---|---|
| Primary Hepatocytes | Study hepatic metabolism and toxicity | IVIVE, drug clearance prediction [65] |
| Human Liver Microsomes | Phase I metabolism studies | Metabolic stability assessment [65] |
| Open TG-GATEs Database | Toxicogenomics reference data | Model training and validation [66] |
| Zebrafish Embryos | Vertebrate model not classified as experimental animals | Bridging in vitro and in vivo testing [63] |
| S1500+ Gene Set | Toxicity-relevant gene panel | Transcriptomic analysis in toxicology [66] |
| Biphasic Dissolution System | Simulate dissolution and absorption | IVIVC for BCS Class II drugs [67] |
Emerging Solutions and Technological Advances
Artificial Intelligence and Advanced Modeling
Novel computational approaches are showing promise in bridging the correlation gap:
AIVIVE Framework: This AI-powered approach integrates generative adversarial networks (GANs) with local optimizers guided by biologically relevant gene modules to improve prediction accuracy [66]. The framework demonstrated high overlap with differentially expressed genes, including Cytochrome P450 enzymes often underrepresented in vitro.
Structure–Tissue Exposure/Selectivity–Activity Relationship (STAR): This proposed classification improves upon traditional structure-activity relationship (SAR) by incorporating tissue exposure and selectivity [60]. It categorizes drug candidates into four classes based on potency/specificity and tissue exposure/selectivity to better predict clinical success.
Level A, B, and C Correlations: These statistical approaches establish relationships between in vitro dissolution profiles and in vivo performance, with Level A representing the highest correlation level involving point-to-point relationships [68].
The following diagram illustrates the AIVIVE framework workflow for enhancing in vitro to in vivo extrapolation:
Improved Model Systems and Testing Strategies
Integrated Testing Cascades: Developing validated, reliable, and predictive in vitro assay cascades that can be used early in preclinical development [64]. Such cascades should establish firm correlation between in vitro assays and their in vivo counterparts.
Zebrafish as a Bridge Model: Zebrafish embryos offer a unique compromise, providing in vivo complexity while being cost-effective and not classified as experimental animals until 5-6 days post-fertilization [63]. This model supports the 3Rs principles (Replacement, Reduction, and Refinement) in research.
Mechanistic Pharmacokinetic Modeling: These approaches assist in understanding drug behavior in the body and play an essential role in new drug development [67]. Establishing IVIVC bridges the gap between in vitro characteristics and in vivo drug behaviors, providing valuable insights for formulation development.
The correlation crisis between in vitro and in vivo results remains a significant challenge in biomedical research and drug development. The fundamental biological differences between isolated systems and whole organisms, combined with methodological limitations in current testing approaches, contribute to the high failure rate of drug candidates in clinical trials. However, emerging technologies—particularly AI-enhanced prediction models, improved testing cascades, and better integrated data analysis—offer promising pathways to enhance translational success.
Addressing these challenges requires acknowledging the limitations of reductionist approaches while systematically working to identify where in vitro systems provide meaningful predictive value. By adopting more sophisticated models, implementing advanced computational approaches, and maintaining rigorous correlation standards, researchers can gradually improve the predictive power of preclinical studies, ultimately enhancing drug development efficiency and success.
Biomedical research is undergoing a fundamental transformation, moving away from traditional models that poorly predict human outcomes toward advanced in vitro systems that more accurately recapitulate human physiology and pathology. The notoriously high failure rates of the current drug development process, largely attributable to the limitations of animal models and conventional two-dimensional (2D) cell cultures, have driven this shift [69]. Traditional 2D cultures, while useful for basic research, fail to replicate the rich microenvironment and complex cellular interactions observed in living tissues [70] [71]. Similarly, animal models suffer from significant interspecies differences that limit their predictive value for human disease outcomes [69]. These challenges have catalyzed the development of advanced in vitro models, particularly those utilizing three-dimensional (3D) architectures, multiple cell types in co-culture, and human primary cells, which collectively bridge the translational gap between basic research and clinical success.
These advanced models represent a continuum of increasing physiological complexity, moving from simple monocultures to systems that incorporate human-relevant cell types, structural dimensionality, and the multicellular interactions that define living organs. The integration of 3D co-culture systems with human primary cells has emerged as a particularly promising approach for disease modeling, drug screening, and personalized medicine applications [72] [73]. This guide provides a comprehensive comparison of these advanced models against traditional alternatives, supported by experimental data and detailed methodologies, to inform researchers and drug development professionals about their capabilities, applications, and performance advantages in the context of biomaterial testing and therapeutic development.
Model Evolution: From 2D Simplicity to 3D Complexity
Fundamental Limitations of Traditional 2D Models
The conventional 2D cell culture system has served as a fundamental tool in biological research for decades, but its simplicity comes with significant drawbacks. In these monolayer systems, cells are grown on flat, rigid plastic or glass surfaces, which fails to reproduce the physiological reality of natural tissue environments [70]. This artificial environment leads to altered cell morphology, polarity, and behavior, ultimately resulting in data that does not faithfully reproduce in vivo responses [70]. The loss of defined tissue organization and proper cell-to-cell and cell-to-matrix interactions in 2D cultures fundamentally limits their translational relevance [70]. Furthermore, genes promoting undesired cell proliferation can be artificially enhanced in 2D systems, leading to cellular behaviors that diverge from those observed in human patients [71].
The Rise of Three-Dimensional Culture Systems
Three-dimensional culture systems overcome these limitations by simulating the physiological context of an organism, from the molecular level to cellular, tissue, and organ complexity levels [70]. The complex organization of living bodies can be better reproduced in 3D models, which incorporate multiple cell types and their interactions, including cell/cell and cell/matrix connections, as well as interactions with their external environment [70]. Cells cultured in 3D environments more closely mimic in vivo cell behavior and organization—both morphologically and physiologically—by recreating their natural microenvironment and facilitating proper cell differentiation and tissue organization [71]. These advantages make 3D systems invaluable for modeling human biology and disease states with higher fidelity.
Table 1: Comparison of 2D, 3D, and Advanced 3D Co-culture Model Characteristics
| Feature | Traditional 2D Models | Simple 3D Models | Advanced 3D Co-cultures with Primary Cells |
|---|---|---|---|
| Spatial Architecture | Flat, monolayer | Three-dimensional structure | Physiologically relevant 3D tissue organization |
| Cell-Cell Interactions | Limited to peripheral contact | Enhanced through spontaneous aggregation | Controlled, tissue-specific interactions |
| Cell-ECM Interactions | Artificial, on rigid plastic | Biomimetic, but often simplified | Native-like with complex ECM composition |
| Physiological Relevance | Low, altered gene expression | Moderate, better mimicry of tissue morphology | High, recapitulates key aspects of human tissue |
| Predictive Value for Drug Responses | Poor, high failure rate in translation | Improved, especially for cancer research | High, demonstrated success in disease modeling |
| Cost & Technical Complexity | Low, well-established | Moderate, requires specialized matrices | High, requires multiple cell types and optimization |
| Applications | Basic research, high-throughput screening | Toxicology, basic disease mechanisms | Personalized medicine, complex disease modeling, regenerative medicine |
Complex In Vitro Models (CIVMs) and Co-culture Systems
The most advanced category of these systems has been termed Complex In Vitro Models (CIVMs), defined as systems in a 3D multicellular environment within a biopolymer or tissue-derived matrix that incorporates primary or stem cell-derived cells, immune system components, and mechanical factors such as stretch or perfusion, or at least two of these elements [6]. These models seek to reconstruct the organ- or tissue-specific characteristics of the extracellular microenvironment, allowing for enhanced physiological correlation of cultured cells and better mimicking of in vivo conditions without the ethical concerns associated with animal experimentation [6]. A prominent example is the development of 3D primary co-culture models of the human airway, which consist of human primary cells representing a differentiated mucociliary airway epithelium, an underlying submucosa with fibroblasts, and an endothelial interface [72]. Such systems demonstrate how increasing cellular complexity within a 3D architecture creates more physiologically relevant models for research and drug testing.
Experimental Approaches and Methodologies
3D Culture Techniques: Scaffold-Based and Scaffold-Free Systems
The engineering of advanced 3D models primarily utilizes two complementary approaches: scaffold-based and scaffold-free systems. Each offers distinct advantages and is suited to different research applications and tissue types.
Scaffold-Based Systems utilize supportive matrices that provide structural support and biochemical cues that mimic the native extracellular matrix (ECM). These systems dominated the market with 48.85% of revenue in 2024 [74] and can be further categorized by material composition:
- Natural Hydrogels: Including collagen, Matrigel, alginate, gelatin, hyaluronic acid, agarose, laminin, or fibrin [71]. These materials are biodegradable, bioactive, and provide natural integrin-binding sites and growth factors that regulate cell behavior through activating signaling cascades [71]. However, they may exhibit poor mechanical properties and batch-to-batch variability.
- Synthetic Polymers: Including polyester, polyethylene glycol (PEG), polyamide, or polylactic acid (PLA) [71]. These offer higher consistency, reproducibility, and customization properties than natural hydrogels but often lack cell recognition sites [71].
- Composite Materials: Combining natural and synthetic materials to optimize both biological and mechanical properties. For example, adding ceramic materials like hydroxyapatite (HA) and β-tri-calcium phosphate (TCP) to polymeric polycaprolactone (PCL) scaffolds has been shown to enhance mechanical properties and cell proliferation rate [71].
Scaffold-Free Systems generate 3D cellular aggregates without supporting matrices, relying on cells' innate ability to self-assemble. These systems are growing at the fastest compound annual growth rate (CAGR of 9.1%) in the 3D cell culture market [74] and include several technical approaches:
- Forced-Floating Methods: Using low-adhesion polymer-coated well plates where spheroids form after centrifugation [71].
- Hanging Drop Methods: Allowing cell suspension aliquots to aggregate in droplets, enabling precise control over spheroid size [71].
- Agitation-Based Approaches: Using rotating bioreactors to create microgravity conditions that prevent adhesion to container walls and promote aggregate formation [71].
Diagram 1: 3D Culture System Selection Workflow
Protocol: Establishing a 3D Primary Co-culture Model of the Human Airway
The development of advanced co-culture models requires careful attention to cell sourcing, matrix composition, and culture conditions. The following protocol outlines the key steps for establishing a 3D primary co-culture model of the human airway, based on published methodology [72]:
Cell Sourcing and Isolation: Obtain human primary bronchial epithelial cells from patient biopsies or organ transplant donors. Isolate fibroblasts from the underlying submucosa and endothelial cells from vascular tissues.
Gradual Complexity Increase: Begin with an established air-liquid interface (ALI) culture system and progressively increase complexity by sequentially adding different primary cell types.
Matrix and Coating Optimization: Test different coatings and extracellular matrix compositions to identify the optimal substrate for supporting multicellular organization. Natural hydrogels like collagen or laminin are often preferred for their bioactive properties.
Three-Dimensional Matrix Incorporation: Embed cells within a three-dimensional matrix that allows for proper spatial organization and cell-cell interactions. A combination of collagen and Matrigel often provides suitable mechanical and biochemical properties.
Differentiation and Maturation: Culture the assembled construct under air-liquid interface conditions for 4-6 weeks to promote differentiation into a mucociliary airway epithelium with underlying connective tissue and endothelial components.
Validation and Characterization: Assess model fidelity through histology (showing pseudostratified epithelium with ciliated cells and goblet cells), immunohistochemistry (for cell-type-specific markers), and functional assays (such as mucociliary clearance and barrier function measurements).
This protocol results in a reproducible 3D in vitro model of the airway consisting of human primary cells representing a differentiated mucociliary airway epithelium, an underlying submucosa with fibroblasts, and an endothelial interface [72].
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful implementation of advanced 3D co-culture models requires specific reagents and materials carefully selected for their functional properties.
Table 2: Essential Research Reagents for Advanced 3D Co-Culture Models
| Reagent/Material | Function | Examples & Applications |
|---|---|---|
| Natural Hydrogels | Mimics native ECM, provides biochemical cues | Collagen for stromal support; Matrigel for epithelial morphogenesis |
| Synthetic Polymers | Provides structural support with controlled properties | PEG for customizable mechanical properties; PLA for biodegradable scaffolds |
| Cell Culture Media | Supports growth and differentiation of multiple cell types | Specialized media formulations for stem cell differentiation; co-culture media |
| Primary Human Cells | Provides human-relevant physiology and patient-specific responses | Bronchial epithelial cells for airway models; hepatocytes for liver models |
| Growth Factors & Cytokines | Directs cell differentiation and tissue organization | EGF, FGF, Wnt agonists for stem cell differentiation; tissue-specific morphogens |
| Microfluidic Devices | Enables precise control of microenvironment and perfusion | Organ-on-chip platforms for physiological flow conditions; gradient generation |
Performance Comparison and Validation Data
Predictive Value for Drug Responses and Disease Modeling
Advanced 3D co-culture models with human primary cells have demonstrated superior predictive value across multiple applications, particularly in disease modeling and drug development. The enhanced performance stems from their ability to more accurately recapitulate human physiology and pathological processes compared to traditional models.
In pharmaceutical research, these models provide a robust platform for drug discovery and development, enabling more accurate predictions of therapeutic efficacy and safety [74]. One compelling example comes from cystic fibrosis research, where human bronchial epithelial cells (HBEs) from patients' lungs have been used to accurately predict efficacy of the CF drugs Kalydeco, Orkambi, and Trikafta [75]. These patient-derived primary cells in 3D culture maintained the actual disease genotype and phenotype, providing a highly accurate in vitro disease model for therapeutic testing [75].
In cancer research, which accounts for 34% of 3D cell culture applications [74], 3D models enable the study of tumor behavior and treatment response in ways that traditional 2D cultures cannot provide. The application of 3D tumor cultures has become particularly valuable for evaluating the pathobiology of human cancer due to their capability of recreating the 3D architecture of tissue [70]. These models replicate tumor heterogeneity and microenvironments, leading to therapies that are more aligned with personalized medicine approaches [70].
Table 3: Comparison of Model Predictive Capacity Across Different Applications
| Application Area | Traditional 2D Models | Animal Models | Advanced 3D Co-cultures with Primary Cells |
|---|---|---|---|
| Drug Efficacy Screening | Poor correlation with clinical outcomes (>75% failure rate) [75] | Moderate, limited by interspecies differences [69] | High, demonstrated accurate prediction of clinical efficacy (e.g., CF drugs) [75] |
| Toxicology Assessment | Limited, misses tissue-specific responses | Moderate, useful for systemic toxicity but limited for human-specific effects | High, enables human-specific toxicity detection in tissue context |
| Cancer Biology Studies | Limited to proliferation studies, lacks tumor microenvironment | Good for tumor growth but limited metastasis prediction | High, recapitulates tumor heterogeneity and drug resistance mechanisms [70] |
| Personalized Medicine | Not applicable | Not applicable | High, patient-derived organoids predict individual drug responses [74] |
| Regenerative Medicine | Limited relevance for tissue formation studies | Useful but limited by immunological and physiological differences | High, enables development of functional tissues for transplantation [74] |
Validation Against Clinical Outcomes: Key Case Studies
The true value of advanced in vitro models is demonstrated through their validation against clinical outcomes. Several key case studies highlight their superior predictive capacity:
Biomaterial Host Response Prediction: A seminal study demonstrated that an in vitro assay characterizing the dynamic inflammatory response of human monocyte-derived macrophages to biomaterials, coupled with in silico analysis (principal component analysis and dynamic network analysis), could accurately predict in vivo remodeling and macrophage polarization responses in a rodent skeletal muscle injury model [7]. The in vitro macrophage response to extracellular matrix (ECM) materials corresponded to constructive remodeling and an increased M2 macrophage presence in vivo, while the response to crosslinked ECM and synthetic materials predicted a foreign body reaction and dominant M1 macrophage response [7]. This approach provides a high-throughput method for predicting host responses to biomaterials without extensive animal testing.
Personalized Oncology Applications: Patient-derived organoids (PDOs) have shown remarkable promise in predicting individual patient responses to cancer therapies. The use of patient-derived primary cells maintains the actual disease genotype and phenotype, enabling more accurate modeling of individual variations in drug response [75]. This approach is particularly valuable in oncology, where tumor heterogeneity and individual variations in drug metabolism significantly impact treatment outcomes.
Regulatory Shifts Supporting Advanced Models: The validation of these advanced approaches has prompted regulatory changes, most notably the "FDA Modernization Act 2.0" passed by the U.S Senate in 2022, which authorized the use of certain alternatives to animal testing, including cell-based assays and computer models, to investigate drug safety and effectiveness [6]. This landmark legislation signals a major shift in that animal tests are no longer indispensable, and complex in vitro models may serve as alternatives for drug safety regulation [6].
Diagram 2: In Vitro to In Vivo Translation Pathway
Current Challenges and Future Directions
Technical and Implementation Barriers
Despite their significant advantages, advanced 3D co-culture models with primary cells face several challenges that limit their widespread adoption. The complexity of analysis represents a significant hurdle, as 3D structures require advanced imaging, multi-omics, or spatial transcriptomics to fully characterize cellular interactions and gradients [70]. These analytical requirements demand specialized expertise, training, and equipment that may not be readily available in all research settings.
Cost considerations also present barriers to implementation. Overall, 3D models are more expensive than 2D models; their assembly may require the purchasing of new laboratory products like hydrogels, scaffolds, and specialized plasticware, as well as equipment, particularly regarding imaging [70]. Scalability remains another critical issue: expanding these models for high-throughput drug screening or population-level studies often requires sophisticated bioreactors and automation technologies that are not yet widely accessible [70].
Standardization and reproducibility challenges further complicate adoption. Currently, robust quality control and standardization measures are essential for reliable and reproducible research results but can be difficult to achieve [70]. Harmonizing protocols for scaffold fabrication, bioprinting, cell sourcing, and analytical readouts remains an ongoing challenge for the field. Best practices for batch-to-batch consistency, sterility, and validated functional assays are critical to minimize variability and enable cross-laboratory comparisons [70].
Emerging Technologies and Future Outlook
Several emerging technologies promise to address these challenges and expand the capabilities of advanced in vitro models. The integration of artificial intelligence (AI) and machine learning (ML) into cell culture processes is set to enhance the efficacy of 3D cultures [74]. By analyzing large datasets, these technologies can improve the design and optimization of culture conditions, leading to better outcomes in drug testing and development [74].
3D bioprinting technologies are advancing rapidly, with companies like CELLINK and CytoNest developing bioprinted tissues for transplantation and drug testing [74]. This approach allows for precise spatial control over cell placement and matrix composition, enabling the creation of more complex and reproducible tissue architectures.
Organ-on-chip technology represents another frontier, projected to grow at a 21.3% CAGR and demonstrating potential to reduce drug development costs by 25% [74]. These microfluidic devices enable precise control over fluid flow and mechanical forces, better replicating the dynamic physiological environment of living organs.
The market outlook reflects strong confidence in these technologies, with the global 3D cell culture market projected to grow at a CAGR of 15% through 2030, driven by demand for alternatives to animal testing, personalized medicine, and drug discovery efficiency [74]. Similarly, the human primary cell culture market is expected to grow from USD 4.10 billion in 2025 to USD 8.61 billion by 2032, exhibiting a compound annual growth rate (CAGR) of 11.2% [73]. This growth trajectory underscores the increasing importance of these advanced models in biomedical research and drug development.
As these technologies mature and overcome current limitations, they are poised to fundamentally transform biomedical research, enabling more human-relevant studies, reducing reliance on animal models, and accelerating the development of safer, more effective therapeutics. The continued refinement of 3D co-culture systems with human primary cells represents a critical pathway toward more predictive, personalized, and successful medical interventions.
The pursuit of reliable biomaterial testing hinges on the ability to create in vitro models that faithfully replicate the complex biological milieu of a living organism. Traditional static cell cultures often fail to predict in vivo performance, leading to high attrition rates in drug development and biomaterial application. This guide objectively compares testing paradigms by examining two critical advancements: the incorporation of blood plasma components and the implementation of dynamic culture conditions. These approaches seek to bridge the gap between the controlled but simplistic in vitro environment ("in glass") and the biologically complex but less controllable in vivo environment ("within the living") [38] [11].
Blood plasma serves as a biologically relevant medium, providing essential proteins, growth factors, and clotting components that mimic the natural extracellular matrix and mediate the host response to biomaterials [76] [77]. Meanwhile, dynamic culture systems introduce biomechanical forces and improve nutrient transport, overcoming the diffusion-limited nature of static cultures and promoting more physiologic cell behaviors [78] [79]. The integration of these elements creates a more predictive testing niche, crucial for researchers, scientists, and drug development professionals aiming to translate in vitro findings into successful clinical outcomes.
The Scientific Foundation: In Vitro vs. In Vivo Testing Paradigms
Defining the Testing Environments
Understanding the fundamental differences between in vitro and in vivo models is essential for interpreting biomaterial testing data.
In Vitro Models: These experiments are conducted outside of living organisms, in controlled laboratory environments such as petri dishes or test tubes [38] [11]. They offer high precision for studying isolated cellular and molecular mechanisms, are cost-effective, and provide faster results than in vivo studies. However, their primary limitation is the inability to replicate the full systemic response of an entire organism, including complex immune interactions and metabolic processes [11].
In Vivo Models: These studies are performed within a living organism, such as animals or humans [38] [11]. They provide critical data on how an entire biological system responds to a biomaterial or drug, offering high physiological relevance. The main drawbacks include ethical concerns, significantly higher costs, longer study durations, and greater complexity in controlling variables [11].
Comparative Framework at a Glance
Table 1: Key Characteristics of In Vitro vs. In Vivo Testing Models
| Aspect | In Vitro Models | In Vivo Models |
|---|---|---|
| Definition | In a controlled lab environment [11] | Within a living organism [11] |
| Complexity | Low; isolated cells or tissues [38] | High; whole-organism systemic response [38] [11] |
| Control | High; variables can be precisely manipulated [11] | Low; complex internal environment is difficult to control [11] |
| Cost & Resources | Lower cost; requires fewer materials [11] | High cost; extensive animal care and monitoring [11] |
| Time to Results | Faster; suitable for high-throughput screening [11] | Longer; involves extended study timelines [11] |
| Ethical Considerations | Lower; no live animals involved [11] | Significant; requires strict ethical oversight [11] |
| Predictive Value | High for mechanistic studies, low for systemic effects [11] | High for whole-body response, crucial for clinical translation [11] |
Blood Plasma as a Bioactive Medium in Biomaterial Testing
Composition and Functional Properties
Blood plasma is an acellular, light-yellow liquid that constitutes about 55% of whole blood. It is a highly complex proteome, comprising approximately 90% water with a rich mixture of proteins, carbohydrates, lipids, salts, enzymes, nutrients, and waste products [76]. Its usefulness as a biomaterial stems from its clotting proteins—primarily fibrinogen and thrombin—which enable the generation of a provisional hydrogel that closely mimics the natural extracellular matrix (ECM) [76]. This fibrin-based matrix is biologically active, containing binding sites for various proteins and growth factors like fibronectin, fibroblast growth factor (FGF), and vascular endothelial growth factor (VEGF), which play an active role in wound healing and cellular interactions [76].
Experimental Data: Plasma-Activated Coagulation Responses
The hemocompatibility of biomaterials is frequently assessed by measuring plasma coagulation responses. A 2023 study systematically evaluated four medical-grade polyurethane biomaterials (Biospan MS/0.4, BioSpan S, BioSpan 2F, and CarboSil 20 80A) for cardiovascular applications using both bovine and human blood plasma [80].
Table 2: Surface Properties and Coagulation Responses of Polyurethane Biomaterials
| Polymer Material | Key Surface Characteristics | Surface Free Energy | Plasma Coagulation Time | FXII Contact Activation |
|---|---|---|---|---|
| Biospan MS/0.4 | Silicone-present surface [80] | Moderate [80] | No significant difference from other PUs [80] | Low, no significant difference [80] |
| BioSpan S | Silicone-present surface [80] | Moderate [80] | No significant difference from other PUs [80] | Low, no significant difference [80] |
| BioSpan 2F | High fluorine content [80] | Lowest [80] | No significant difference from other PUs [80] | Low, no significant difference [80] |
| CarboSil 20 80A | Silicone-present surface [80] | Moderate [80] | No significant difference from other PUs [80] | Low, no significant difference [80] |
| Polystyrene Control | Standard tissue culture plastic [80] | Not Reported | Baseline (shorter) [80] | Not Reported |
The in vitro coagulation assay demonstrated that all polyurethane materials led to improved blood coagulation responses (longer coagulation times) compared to the polystyrene control. Furthermore, a chromogenic assay showed that all PU materials resulted in low Factor XII (FXII) contact activation, with no significant differences among them, consistent with the plasma coagulation data [80]. This suggests that these potential replacement materials exhibit hemocompatibility comparable to the established Biospan MS/0.4.
Experimental Protocol: Plasma Coagulation Testing
Objective: To evaluate the plasma coagulation response and FXII contact activation of biomaterials in vitro [80].
Materials and Reagents:
- Biomaterial Samples: Fabricated as tubes via dipping method on stainless steel rods [80].
- Blood Plasma: Human or bovine blood collected in anticoagulant-containing vacutainers [80].
- Factor XII and Factor XIIa: Commercially obtained for calibration and standard curves [80].
- Chromogenic Substrate (Pefachrome FXIIa/TH 5253): For quantifying FXIIa activity [80].
- Phosphate Buffered Saline (PBS): For dilution and washing steps [80].
Methodology:
- Sample Preparation: Fabricate polymer tubes via multiple dipping cycles onto rods, followed by curing and extraction of residual solvents in deionized water [80].
- Plasma Incubation: Incube the biomaterial tubes with blood plasma under controlled conditions (e.g., 37°C) [80].
- Coagulation Time Measurement: Record the time for a visible clot to form after recalcification or other triggers [80].
- FXII Activation Assay: After contact, incubate plasma with a chromogenic substrate. Measure the rate of color change, which is proportional to the amount of FXIIa generated [80].
- Data Analysis: Compare coagulation times and FXIIa levels between test materials and controls [80].
Dynamic Culture Conditions for Enhanced Physiological Relevance
Principles and System Design
Static culture conditions suffer from limited nutrient diffusion, waste accumulation, and lack of physiologically relevant mechanical stimulation. Dynamic culture systems address these limitations by inducing convective transport through media flow, typically using bioreactors with peristaltic pumps or orbital shakers [78] [79]. These systems enhance nutrient and oxygen distribution to cells throughout a 3D construct, preventing necrosis in the core and promoting more uniform cell distribution and extracellular matrix (ECM) production [78] [79]. Furthermore, they can be designed to apply specific mechanical forces, such as pulsatile flow or shear stress, which are critical for the proper differentiation and function of tissues like blood vessels [78].
Experimental Data: Dynamic vs. Static Culture Outcomes
A study on tissue-engineered vascular grafts (TEVGs) using human cells on electrospun silk fibroin tubular scaffolds directly compared outcomes under dynamic pulsatile flow versus static culture conditions [78].
Table 3: Performance of Tissue-Engineered Vascular Grafts Under Different Culture Conditions
| Performance Metric | Static Culture | Dynamic Culture | Biological Significance |
|---|---|---|---|
| Cell Proliferation | Lower [78] | Enhanced [78] | Faster construct development and cellularity |
| Cell Alignment | Poorly organized [78] | Well-aligned along flow direction [78] | Recapitulates native vessel architecture |
| ECM Production | Lower and less organized [78] | Increased and aligned deposition [78] | Improved mechanical strength and function |
| Phenotype Maintenance | Less stable [78] | Better maintenance of contractile phenotype [78] | Essential for long-term graft functionality |
| Cellular Infiltration | Limited to periphery, heterogeneous [78] | Enhanced depth and uniformity [78] | Prevents core necrosis, enables thicker constructs |
| Endothelial Retention | Poor under shear [78] | Improved with basement membrane mimic [78] | Critical for non-thrombogenic luminal surface |
The results demonstrated that dynamic flow conditions yielded superior outcomes across all measured parameters. The enhanced nutrient and oxygen distribution under convection prevented central necrosis, while the mechanical stimulation from pulsatile flow guided cells to adopt a more natural, aligned morphology and to increase their production of a functional, organized extracellular matrix [78].
Experimental Protocol: Bioreactor Culture for Vascular Grafts
Objective: To develop a tissue-engineered vascular graft under physiological pulsatile flow conditions [78].
Materials and Reagents:
- Tubular Scaffolds: Electrospun silk fibroin scaffolds [78].
- Cell Types: Human Coronary Artery Smooth Muscle Cells (HCASMCs) and Human Aortic Endothelial Cells (HAECs) [78].
- Culture Media: Smooth Muscle Cell Medium (SMCM) and Endothelial Growth Medium-2 (EGM-2) [78].
- Bioreactor System: Consisting of a polycarbonate culture chamber, peristaltic pump, and silicone tubing for gas exchange [78].
- Matrigel: Used as a basement membrane mimic to improve endothelial cell adhesion [78].
Methodology:
- Scaffold Preparation: Fabricate tubular scaffolds via electrospinning, treat with methanol to induce water-insolubility, and sterilize [78].
- Cell Seeding: Seed HCASMCs onto the luminal surface of the scaffold first and allow for adherence. Subsequently, seed HAECs on top, optionally using a Matrigel coating to enhance retention [78].
- Bioreactor Assembly: Mount the cell-seeded scaffold within the bioreactor chamber, connecting it to the flow loop [78].
- Dynamic Culture: Initiate physiological pulsatile flow using the peristaltic pump. Maintain the system in a standard humidified incubator (37°C, 5% CO2) for the culture duration [78].
- Static Control: Culture identical constructs in Petri dishes without flow-induced convection [78].
- Outcome Assessment: Analyze constructs for cell viability, proliferation, alignment, ECM production, and mechanical properties after predetermined time points [78].
Integrated Signaling Pathways in Plasma and Dynamic Culture Environments
The biological response in a sophisticated in vitro niche is governed by complex, interconnected signaling pathways. The diagram below illustrates the key pathway activated by blood plasma contact and the cellular responses modulated by dynamic culture.
Diagram 1: Key signaling pathways in plasma-activated coagulation and dynamic culture mechanotransduction.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successfully implementing these advanced models requires specific reagents and equipment. The following table details key solutions for developing predictive in vitro niches.
Table 4: Essential Research Reagents and Materials for Advanced In Vitro Testing
| Reagent/Material | Function and Application | Specific Examples / Notes |
|---|---|---|
| Blood-Derived Products | Provides a biologically complex medium rich in proteins, clotting factors, and growth factors to mimic the in vivo ECM and signaling environment [76] [77]. | Platelet-Rich Plasma (PRP), Platelet-Poor Plasma (PPP), purified Fibrinogen [77]. Autologous sources avoid immune rejection [76]. |
| Chromogenic Substrates | Enables quantification of specific protease activity (e.g., FXIIa, thrombin) in plasma coagulation assays by producing a colorimetric change upon enzymatic cleavage [80] [81]. | Pefachrome FXIIa/TH 5253 for FXIIa measurement [80]. |
| Medical-Grade Polymers | Serve as the scaffold or biomaterial substrate for testing. Require excellent mechanical properties and initial biocompatibility [80] [82]. | Segmented Polyurethanes (e.g., Biospan, CarboSil) [80], Poly(ε-caprolactone) (PCL) [79], Polylactic acid (PLA) [82]. |
| Bioreactor Systems | Provides dynamic culture conditions through controlled media flow, enabling convective transport of nutrients and application of physiological mechanical stimuli [78]. | Systems with pulsatile flow capability for vascular grafts [78]; orbital shakers for simpler agitation [79]. |
| Electrospun Nanofibrous Scaffolds | Creates a 3D synthetic ECM with high surface-area-to-volume ratio and tunable fiber alignment to guide cell growth and tissue organization [78] [79]. | Silk fibroin [78], PCL [79]. Alignment directs cell orientation and ECM deposition [79]. |
| Factor XII (FXII) | A key zymogen in the contact activation pathway of coagulation; used to study and calibrate plasma-biomaterial interactions [80] [81]. | Commercially obtained from human plasma for in vitro assays [80]. |
The integration of blood plasma components and dynamic culture conditions represents a paradigm shift in biomaterial testing, creating in vitro environments that more accurately predict in vivo performance. Quantitative data demonstrates that plasma provides critical biochemical cues for assessing hemocompatibility, while dynamic systems induce essential biomechanical signals that guide functional tissue development. For researchers in drug development and regenerative medicine, leveraging these advanced models is no longer optional but essential for improving the predictive validity of preclinical studies and accelerating the translation of safe and effective biomaterials and therapies to the clinic.
The Critical Role of Macrophage Polarization in Predicting Fibrotic Responses
Fibrosis, characterized by the excessive accumulation of extracellular matrix (ECM) components, represents a final common pathway for many chronic diseases affecting the heart, liver, kidneys, and lungs [83]. This pathological scarring destroys normal tissue architecture and leads to organ dysfunction, with fibrotic diseases contributing to approximately 45% of all deaths in developed countries [83]. Within this complex pathological process, macrophages have emerged as central regulators due to their remarkable plasticity and ability to influence all stages of fibrogenesis [84] [83]. The polarization state of macrophages—their ability to adopt distinct functional phenotypes in response to microenvironmental cues—serves as a critical determinant of fibrotic progression or resolution [85]. Understanding and predicting these polarization patterns has become essential for evaluating biomaterial safety and developing anti-fibrotic therapies, necessitating robust testing platforms that can accurately model the dynamic interplay between macrophages and their microenvironment [86] [87].
The traditional classification of macrophages into pro-inflammatory (M1) and anti-inflammatory (M2) phenotypes provides a foundational framework, though contemporary research reveals a spectrum of activation states beyond this binary paradigm [84] [83]. M1 macrophages, typically induced by interferon-gamma (IFN-γ) and lipopolysaccharide (LPS), drive initial inflammatory responses through production of reactive oxygen species (ROS) and cytokines including IL-1β, IL-6, and TNF-α [83] [85]. In contrast, M2 macrophages, polarized by IL-4 and IL-13, promote tissue repair and fibrogenesis by producing transforming growth factor-beta (TGF-β) and other mediators that activate fibroblasts and stimulate ECM deposition [88] [83]. The balance between these phenotypes significantly influences whether tissue injury resolves normally or progresses to pathological fibrosis [85].
Comparative Analysis of Testing Platforms for Studying Macrophage-Driven Fibrosis
Evaluating the fibrotic potential of biomaterials and therapeutic compounds requires testing platforms that accurately recapitulate macrophage polarization dynamics. The table below summarizes the key characteristics of current experimental models used in fibrosis research.
Table 1: Comparison of Experimental Platforms for Studying Macrophage Polarization in Fibrosis
| Testing Platform | Key Features | Advantages | Limitations | Primary Applications |
|---|---|---|---|---|
| In Vitro 2D Cultures [86] | Monocultures or cocultures on plastic surfaces; controlled polarization with cytokine cocktails | High throughput; cost-effective; well-established protocols; reduced biological variability | Limited cellular complexity; artificial microenvironment; poor translation to in vivo outcomes | Initial screening of compounds; mechanistic studies of specific pathways |
| Ex Vivo Models (PCLS) [88] | Precision-cut lung slices (PCLS) maintain native tissue architecture and cellular populations | Preserves tissue complexity and cell-cell interactions; more physiologically relevant | Technically challenging; limited viability period; moderate throughput | Investigating macrophage-fibroblast interactions in tissue context; validation studies |
| In Vivo Models [89] | Whole-organism studies (e.g., bleomycin-induced pulmonary fibrosis in mice) | Complete biological system; accounts for systemic immune responses; clinical relevance | High cost; ethical concerns; complex data interpretation; species-specific differences | Preclinical validation of anti-fibrotic therapies; study of disease pathogenesis |
| In Silico Models [86] [87] | Mathematical modeling of macrophage-fibroblast interplay and FBR dynamics | Enables long-term simulations; investigates non-linear interactions and feedback loops; hypothesis generation | Requires experimental validation; limited by parameter availability and model complexity | Predicting long-term FBR outcomes; optimizing biomaterial design parameters |
Each platform offers distinct advantages for investigating macrophage polarization in fibrotic responses. In vitro systems provide controlled environments for mechanistic studies but often fail to capture the complexity of tissue-level responses [86]. Ex vivo models, such as precision-cut lung slices (PCLS), maintain native tissue architecture and have demonstrated utility for investigating profibrotic macrophage polarization in response to IL-4, IL-13, and IL-6 stimulation [88]. In vivo models remain the gold standard for preclinical validation, as exemplified by studies showing that combination therapy with nintedanib and pexidartinib (PLX3397) can reverse established fibrosis by modulating macrophage polarization [89]. Emerging in silico approaches integrate data from these experimental systems to create predictive models of the foreign body response (FBR), incorporating key material properties such as stiffness and immunogenicity that influence macrophage behavior [86].
Experimental Methodologies for Assessing Macrophage Polarization
Establishing Profibrotic Macrophage Polarization Ex Vivo
The PCLS model provides a representative methodology for investigating macrophage polarization in a complex, tissue-relevant environment. The experimental workflow involves several key stages [88]:
- Tissue Preparation: Precision-cut lung slices (approximately 300 µm thickness) are prepared from murine lung tissue using a vibratome and maintained in serum-free culture medium.
- Profibrotic Polarization Induction: Slices are treated with a polarization cocktail (PC) consisting of IL-4 (20 ng/mL), IL-13 (20 ng/mL), and IL-6 (20 ng/mL) over a 72-hour time course. This cytokine combination drives macrophages toward a profibrotic phenotype.
- Viability Assessment: Tissue viability and architecture are confirmed through visual inspection and metabolic assays to ensure the polarization protocol does not induce adverse effects.
- Phenotype Characterization: Polarized macrophages are assessed using multiple markers: Arginase-1 (Arg1), CD206, YM1, and CCL17 through immunohistochemistry and quantitative PCR.
- Model Validation: Macrophage depletion using clodronate treatment confirms the specific contribution of macrophages to the observed profibrotic responses.
This approach enables researchers to investigate macrophage polarization within the context of native tissue architecture, bypassing the artificial recapitulation of the lung milieu required in simpler in vitro systems [88].
Computational Modeling of Macrophage-Fibroblast Interplay in Fibrosis
Mathematical modeling provides a complementary approach to experimental systems for understanding fibrosis pathogenesis. A representative methodology for constructing such models includes [87]:
- Transcriptomic Data Collection: RNA sequencing of macrophages and fibroblasts under 44 distinct experimental conditions representing varied immune and metabolic contexts (e.g., pro-inflammatory, hypoxic, coculture).
- Pathway Analysis: Single-sample Gene Set Enrichment Analysis (ssGSEA) to identify major drivers of differential gene expression, including hypoxia, ROS pathway, glycolysis, and TGF-β signaling.
- Multi-level Computational Analysis:
- First-level: Analysis of individual variables (immune stimulation, hypoxia, coculture)
- Second-level: Analysis of combinatorial effects of two variables
- Third-level: Analysis of three-variable interactions
- Model Construction: Development of ordinary differential equation-based models that incorporate key cellular interactions and feedback loops.
- Validation: Comparison of model predictions with experimental data from human tissue samples (e.g., renal allografts with fibrosis) to assess predictive capacity.
This integrated experimental-computational approach has identified distinct fibrotic states ("hot" and "cold" fibrosis) characterized by different cellularity patterns and has demonstrated how specific combinations of metabolic and inflammatory cues drive irreversible fibrosis development [87].
Signaling Pathways in Macrophage Polarization and Fibrosis
The differentiation of macrophages into profibrotic phenotypes is regulated by complex signaling networks that respond to both biochemical and mechanical cues. The diagram below illustrates the key pathways involved in macrophage polarization and their contribution to fibrotic responses.
Diagram 1: Signaling pathways governing macrophage polarization in fibrosis. M1 macrophages drive initial inflammation, while M2 macrophages promote fibrogenesis through TGF-β and other mediators.
The polarization process involves multiple coordinated signaling events. CSF1R signaling plays a particularly crucial role in determining macrophage phenotype, with inhibition of this pathway demonstrating therapeutic potential in pulmonary fibrosis models [89]. The TGF-β/Smad pathway represents another central signaling axis in fibrosis, inducing transcription of profibrotic genes in both Smad-dependent and independent manners [83]. Additional pathways including Wnt/β-catenin, Notch, and MAPK signaling further contribute to the regulation of macrophage function and fibrotic responses [83]. These signaling networks integrate inputs from the tissue microenvironment, including biochemical cues (cytokines, growth factors) and mechanical signals (matrix stiffness), to determine macrophage polarization states and subsequent fibrotic outcomes [86] [85].
The Scientist's Toolkit: Essential Reagents for Macrophage Polarization Research
Table 2: Key Research Reagents for Studying Macrophage Polarization in Fibrosis
| Reagent Category | Specific Examples | Function in Research | Experimental Applications |
|---|---|---|---|
| Polarization Cytokines [88] [85] | IL-4, IL-13, IL-6, IFN-γ, LPS | Induce specific macrophage phenotypes; M2 polarization (IL-4, IL-13) and M1 polarization (IFN-γ, LPS) | In vitro and ex vivo polarization protocols; concentration-dependent studies |
| Macrophage Markers [88] [84] | Arginase-1 (Arg1), CD206, YM1, iNOS, CD86 | Identify and quantify macrophage polarization states; M2 markers (Arg1, CD206) and M1 markers (iNOS, CD86) | Immunohistochemistry, flow cytometry, RNA expression analysis |
| Signaling Inhibitors [89] | Pexidartinib (PLX3397), Nintedanib | Target specific pathways (CSF1R, tyrosine kinases) to modulate macrophage polarization and fibrotic responses | Mechanistic studies; therapeutic intervention experiments |
| Viability Assays [30] | MTT, ATP-based luminescence, dye exclusion tests (trypan blue) | Assess cell health and function; confirm polarization conditions do not adversely affect viability | Quality control for ex vivo models; cytotoxicity screening |
| Fibrosis Markers [88] [83] | α-SMA, Collagen I, Fibronectin, FSP-1 | Evaluate fibroblast activation and ECM deposition as downstream effects of macrophage polarization | Histological assessment; protein and gene expression analysis |
This toolkit enables researchers to precisely manipulate, characterize, and evaluate macrophage polarization states and their functional consequences in fibrotic contexts. The selection of appropriate reagents and assays depends on the specific research questions and experimental models being employed. For example, polarization cytokines allow controlled induction of specific macrophage phenotypes, while signaling inhibitors facilitate mechanistic studies by disrupting specific pathways [88] [89]. Comprehensive assessment typically requires multiple macrophage markers to accurately characterize polarization states, as single markers may not capture the full complexity of macrophage phenotypes, especially given the limitations of the traditional M1/M2 classification scheme [84].
The critical role of macrophage polarization in fibrotic responses underscores the importance of employing complementary testing platforms that capture different aspects of this complex biological process. While in vitro systems offer control and throughput for initial screening, and ex vivo models provide tissue-level context, in vivo studies remain essential for validating findings in physiologically relevant environments [88] [86] [89]. The emerging integration of computational modeling with experimental data represents a promising approach for predicting long-term outcomes and optimizing therapeutic interventions [86] [87].
Accurate prediction of fibrotic responses requires careful consideration of macrophage polarization dynamics across these platforms. The polarization state of macrophages serves not only as a marker of fibrotic progression but also as a potential therapeutic target, as demonstrated by combination therapies that successfully reverse established fibrosis by modulating macrophage phenotypes [89]. As research continues to reveal the complexity of macrophage biology beyond the traditional M1/M2 dichotomy, testing platforms must evolve to capture this heterogeneity and its functional consequences in fibrotic diseases [84] [83]. Through the strategic integration of complementary testing approaches, researchers can enhance the predictive accuracy of fibrotic responses and accelerate the development of effective anti-fibrotic therapies.
Integrating Computational Modeling and Machine Learning for Enhanced Prediction
The field of biomaterial testing is undergoing a transformative shift, moving away from traditional, resource-intensive "trial-and-error" methods toward an integrated computational approach [90]. This evolution is critical for bridging the persistent gap between in vitro laboratory results and in vivo clinical outcomes. The emerging paradigm combines two powerful methodologies: mechanistic computational modeling, rooted in first principles of physics and biology, and data-driven machine learning, which excels at finding complex patterns in large datasets [91] [92]. This guide provides a structured comparison of these approaches, their synergistic integration, and their application for enhancing prediction in biomaterial research and development.
The fundamental challenge in biomaterial science lies in the inherent limitations of both in vitro and in vivo testing. In vitro models, while controlled and reproducible, often fail to capture the complexity of living systems. In vivo models, though more physiologically relevant, are expensive, time-consuming, and raise ethical concerns [93]. Computational modeling and machine learning offer a complementary "in silico" approach that can enhance predictive accuracy, reduce experimental burden, and accelerate the development timeline [91] [90].
Theoretical Foundations: Mechanistic Modeling vs. Machine Learning
Core Principles and Comparative Strengths
The table below outlines the fundamental characteristics of mechanistic modeling and machine learning approaches, highlighting their complementary nature in biomaterial prediction.
Table 1: Fundamental Characteristics of Modeling Approaches
| Feature | Mechanistic Modeling | Machine Learning (ML) |
|---|---|---|
| Foundation | Based on first principles (physics, biology) [91] | Learns patterns from data [92] |
| Data Requirements | Can operate with sparse data [92] | Requires large, high-quality datasets [90] |
| Output | Predicts system behavior and explains underlying mechanisms [91] | Predicts outcomes based on learned correlations [90] |
| Interpretability | High (transparent causality) [91] | Often low ("black box") [90] |
| Generalization | Strong generalizability due to physical basis [91] | May struggle with extrapolation beyond training data [92] |
| Primary Use Case | Hypothesis testing, understanding cause-effect relationships [91] | Pattern recognition, prediction from complex datasets [93] |
The Integrationist Framework
The dichotomy between these approaches is increasingly seen as artificial. The most powerful predictive frameworks strategically combine them [91] [92]. Mechanistic models provide a structured, knowledge-based skeleton that respects biological and physical constraints, while machine learning enhances these models by calibrating them to real-world observational data and identifying patterns that may not be evident from first principles alone [94]. This synergy is akin to navigating between the "mountain view" (integrative, top-down) and the "village view" (reductive, bottom-up) of biological systems [91].
Application in Biomaterial Testing: From In Vitro to In Vivo Prediction
Predicting Osteogenic Differentiation of Stem Cells
A critical application in bone tissue engineering is predicting the osteogenic differentiation of Mesenchymal Stem Cells (MSCs). Traditional methods rely on endpoint detection (e.g., staining), which is destructive and provides lagging indicators [93]. Machine learning models have been successfully applied to achieve early, non-invasive prediction.
Table 2: Machine Learning Models for Predicting MSC Osteogenic Differentiation
| ML Model | Application Context | Reported Performance | Advantages | Limitations |
|---|---|---|---|---|
| ResNet-50 | Prediction using cellular morphology images from live-cell imaging [93] | AUC > 0.96, Accuracy > 96.3% at 24 hours [93] | Residual blocks prevent vanishing gradient; suitable for high-res images [93] | Requires large datasets (≥10,000 images); sensitive to imaging parameters [93] |
| VGG19 | Morphology-based prediction of adipogenic/osteogenic differentiation [93] | Lower performance than ResNet-50 [93] | - | Overfits on small datasets; high computational cost [93] |
| Vision Transformers (ViTs) | Medical image recognition for cell morphology [93] | Promising for global feature capture [93] | Self-attention mechanism captures global features better than CNNs [93] | Emerging application, performance under evaluation [93] |
| LASSO | Omics data analysis for biomarker discovery [93] | Not specified | Feature selection for high-dimensional data [93] | Linear model, may miss complex interactions [93] |
| Random Forests | Omics data analysis and biomaterial screening [93] | Not specified | Handles non-linear relationships; robust to outliers [93] | Less interpretable than simpler models [93] |
Experimental Protocol: Morphology-Based Prediction of Osteogenesis
Objective: To non-invasively predict the osteogenic differentiation potential of MSCs using bright-field images and a ResNet-50 deep learning model [93].
Methodology:
- Cell Culture & Imaging: Plate human MSCs and initiate osteogenic induction. Acquire bright-field images at multiple time points (e.g., days 0, 1, 3, 5, and 7) using phase-contrast microscopy [93].
- Data Labeling: Correlate images with endpoint differentiation confirmation using traditional methods (e.g., Alizarin Red S staining, ALP activity) [93].
- Feature Extraction: The ResNet-50 model automatically extracts relevant morphological features (e.g., cell area, edge roughness, texture) from the images [93].
- Model Training & Validation: Train the ResNet-50 model on a large dataset of images (≥10,000 recommended). Use a separate validation set to assess prediction accuracy, AUC, sensitivity, and specificity. Performance is often compared against traditional biochemical assays [93].
Design and Optimization of Biomaterials
The "trial-and-error" approach in biomaterials development leads to substantial waste of resources [90]. AI and ML are now being deployed to predict material properties, optimize compositions, and design novel materials.
Case Study: Magnesium Alloys for Biodegradable Implants
Magnesium (Mg) alloys are promising for biodegradable implants but face challenges with corrosion resistance and complex deformation behavior [95]. ML models are used to navigate the high-dimensional design space of alloy compositions and processing parameters.
Table 3: AI/ML Applications in Mg Alloy Development for Biomedicine
| AI/ML Model | Application in Mg Alloys | Function |
|---|---|---|
| Artificial Neural Networks (ANNs) | Modeling mechanical behavior (yield strength, tensile strength) [95] | Establish non-linear relationships between composition/processing and properties [95] |
| Convolutional Neural Networks (CNNs) | Microstructure analysis from images (grain size, texture) [95] | Link microstructural features to performance metrics [95] |
| Random Forests (RF) & Support Vector Regression (SVR) | Corrosion modeling and degradation prediction [95] | Predict biodegradation rates in physiological environments [95] |
| Generative Adversarial Networks (GANs) & Bayesian Optimization | AI-driven alloy design and inverse design [95] | Generate novel alloy compositions with desired properties [95] |
Experimental Protocol: Data-Driven Design of Polymer Excipients
Objective: To employ machine learning for the inverse design of novel polymer excipients for pharmaceutical amorphous solid dispersions [96].
Methodology:
- Dataset Curation: Compile a comprehensive dataset of known polymer structures and their key properties relevant to drug formulation (e.g., glass transition temperature, solubility parameters, drug-polymer miscibility) [96].
- Model Selection and Training: Implement a machine learning model (e.g., a neural network or random forest) to learn the complex relationships between chemical descriptors of polymers and their target properties [96] [90].
- Inverse Design and Generation: Use the trained model in reverse: input the desired performance criteria to generate candidate polymer structures that are predicted to meet these specifications [95].
- Synthesis and Validation: Synthesize the top-predicted candidates and experimentally validate their performance in vitro, creating a feedback loop to refine the ML model [96].
Integrated Workflows for Enhanced Prediction
The true power of computational prediction is realized when mechanistic and data-driven models are integrated into a cohesive workflow. This is particularly valuable for translating in vitro results to in vivo outcomes, a multi-scale problem.
This integrated workflow allows researchers to:
- Calibrate Multi-scale Models: Use ML to parameterize mechanistic models (e.g., cardiac electrophysiology models spanning from ion channels to tissue-level excitation) with in vitro data [91].
- Account for Variability: Create virtual populations or cohorts of models that reflect inter-individual variability, moving beyond the "average human" model to enable in silico clinical trials [91].
- Generate Digital Twins: Develop patient-specific models that can be updated with clinical data to predict individual treatment outcomes, a key step in personalized medicine [91].
Successful implementation of integrated modeling requires a suite of computational and data resources.
Table 4: Essential Reagents and Resources for Integrated Computational Research
| Category | Item | Function/Description |
|---|---|---|
| Computational Models | Artificial Neural Networks (ANNs) / Deep Neural Networks [90] [95] | Core ML architecture for learning complex, non-linear relationships from data. |
| Convolutional Neural Networks (CNNs) [93] [95] | Specialized for image-based data (e.g., cell morphology, microstructure). | |
| Random Forests / Gradient Boosting Machines [93] [95] | For tabular data analysis, classification, and regression tasks. | |
| Data Resources | Multi-omics Repositories (Transcriptomics, Proteomics, Metabolomics) [92] [93] | Provide large-scale biological data for training and validating ML models. |
| Clinical & Public Health Data Repositories [92] | Source of real-world evidence for model validation and understanding population variability. | |
| Standardized Biomaterial Datasets [97] | Curated data on material properties and biological responses, essential for data-driven design. | |
| Software & Platforms | Open-Source Analysis Tools & Software Platforms (e.g., Python, R, TensorFlow, PyTorch) [92] | Foundational programming environments and libraries for building and training models. |
| Vetted Computational Pipelines [92] | Standardized workflows for data preprocessing, model training, and validation to ensure reproducibility. | |
| Knowledge Bases | Physics-Based Simulation Software (e.g., for Finite Element Analysis, Molecular Dynamics) [95] | Provide mechanistic modeling capabilities that can be integrated with ML. |
| Systems Biology Models & Databases (e.g., Physiome Project) [91] [94] | Provide established, peer-reviewed mechanistic frameworks for building larger multi-scale models. |
The integration of computational modeling and machine learning is no longer a speculative future for biomaterial science but a present-day necessity for enhancing predictive accuracy. As summarized in this guide, mechanistic models provide the causal, physiological framework, while machine learning brings the power to discern complex patterns from high-dimensional data. Their synergy offers a robust in silico toolkit to better translate in vitro findings into successful in vivo outcomes, ultimately accelerating the development of safer and more effective biomaterials and therapies. Success in this endeavor hinges on continued progress in data standardization, model interpretability, and, most importantly, a community-wide commitment to collaborative, reproducible, and open science [92] [94].
Making the Final Call: A Comparative Framework for Data Integration and Decision-Making
In the field of biomaterial testing and drug development, the choice between in vitro (in glass) and in vivo (within the living) methodologies is not merely a matter of preference but a fundamental strategic decision. In vitro studies are conducted outside a living organism, typically in controlled laboratory environments like petri dishes or test tubes, allowing researchers to examine microorganisms or human and animal cells in culture [2]. In contrast, in vivo studies are performed in or on a whole living organism, such as animals or humans, providing insight into complex biological interactions within their natural environment [2] [11]. While each approach offers distinct advantages, relying exclusively on one creates significant blind spots in safety and efficacy profiling. A comprehensive analysis of experimental data and regulatory frameworks reveals that an integrated methodology is not merely beneficial but non-negotiable for advancing reliable medical technologies.
Section 1: Direct Comparison of In Vitro and In Vivo Approaches
Fundamental Characteristics and Applications
The following table outlines the core distinctions between these two foundational methodologies:
| Aspect | In Vitro Models | In Vivo Models |
|---|---|---|
| Definition | Research conducted outside a living organism, in a controlled lab environment [11] [45] | Research conducted within a whole, living organism [11] [45] |
| Scope of Study | Focused on isolated cells, tissues, or molecular components [2] [11] | Studies the entire organism, providing holistic, systemic data [11] |
| Accuracy & Relevance | Precise for cellular mechanisms but limited in predicting whole-organism response [11] [23] | Highly clinically relevant for predicting effects in humans [11] |
| Cost & Resources | Cost-effective; requires fewer materials and equipment [11] | High cost due to animal care, monitoring, and extensive resources [11] |
| Ethical Considerations | Lower ethical concerns; no live animals involved [11] | Significant ethical concerns, especially with animal testing [21] [11] |
| Time to Results | Faster, more focused experiments ideal for early-stage research [11] | Longer duration due to complex study requirements and monitoring [11] |
| Primary Applications | Early-stage drug screening, mechanistic studies, personalized medicine [2] [11] | Drug discovery, toxicology studies, complex disease modeling [2] [11] |
Quantitative Data from Comparative Studies
Empirical evidence consistently demonstrates the performance gap between isolated systems and whole organisms. The table below summarizes key findings from a comparative study of biomaterials used for orthopedic soft tissue augmentation:
| Biomaterial Tested | In Vitro Cell Viability & Retention | In Vivo Tissue Healing & Regeneration |
|---|---|---|
| Cross-Linked Porcine Small Intestine (CLPSI) | Consistently superior [98] | Favorable properties for clinical application [98] |
| Non-Cross-Linked Porcine Small Intestine (NCLPSI) | Superior cell viability, retention, and infiltration [98] | Replaced with regenerative tissue most rapidly; scored highest in ease of use [98] |
| Non-Cross-Linked Human Dermis | Allowed retention of viable cells, but was not superior [98] | Data not specified in abstract, implied less rapid replacement [98] |
| Cross-Linked Porcine Dermis | Allowed retention of viable cells, but was not superior [98] | Data not specified in abstract, implied less rapid replacement [98] |
| Non-Cross-Linked Fetal Bovine Dermis | Allowed retention of viable cells, but was not superior [98] | Data not specified in abstract, implied less rapid replacement [98] |
This data underscores a critical point: materials that perform excellently in controlled in vitro environments do not always translate to equivalent in vivo outcomes. The non-cross-linked porcine small intestine (NCLPSI),
which demonstrated superior cell infiltration in vitro, also showed the most rapid replacement with regenerative tissue in vivo, highlighting a rare but valuable concordance [98]. Furthermore, a major limitation of in vitro models is their frequent use of cancerous cell lines, which have substantially abnormal functions, and the general absence of biokinetics, which can lead to misinterpretation of data [23].
Section 2: Experimental Protocols in Biocompatibility and Biomaterial Testing
The "Big Three" Biocompatibility Tests
For medical devices, the ISO 10993 standard mandates a series of biocompatibility tests, with the "Big Three"—cytotoxicity, irritation, and sensitization—being central to nearly all device evaluations [21].
Diagram 1: The "Big Three" Biocompatibility Testing Workflow
Detailed Protocol: Cytotoxicity Testing (ISO 10993-5:2009)
Objective: To assess whether a medical device's materials or components can cause harm to living cells [21].
- Test Sample Preparation: Medical devices are tested as extracts. The device or its components are immersed in extraction solvents (e.g., physiological saline, vegetable oil, cell culture medium) under specified conditions [21].
- Cell Lines: Commonly used mammalian cell lines include Balb 3T3 (fibroblasts), L929 (fibroblasts), and Vero (kidney-derived epithelial cells) [21].
- Exposure: Cultured cells are exposed to the device extracts for approximately 24 hours [21].
- Evaluation Endpoints:
- Cell Viability: Measured using assays like MTT, XTT, or Neutral Red Uptake, which quantify metabolic activity [21].
- Morphological Changes: Microscopic observation of changes in cell shape or structure [21].
- Cell Detachment and Lysis: Assessment of cells detaching from the culture substrate or undergoing membrane damage [21].
- Acceptance Criteria: While ISO 10993-5 does not define strict universal criteria, it provides guidance for data interpretation. Typically, cell survival of 70% and above (when testing neat extract) is considered a positive sign of low cytotoxicity [21].
Mechanical Testing Protocols for Biomaterials
Characterizing the mechanical properties of biomaterials is crucial for applications in disease diagnosis, tissue modeling, and engineering. The table below details three common methods, as demonstrated in a comparative study on silicone elastomers and polyacrylamide hydrogels [99]:
| Testing Method | Protocol Summary | Key Metrics | Applicable Materials |
|---|---|---|---|
| Pipette Aspiration (PA) | A micropipette applies suction pressure to the material surface, and the length of the aspirated segment is measured [99]. | Aspiration length vs. pressure used to calculate an effective elastic modulus [99]. | Soft tissues, hydrogels, single cells; ideal for small, fragile samples [99]. |
| Nanoindentation (NI) | A tip (often spherical, 100–1000 μm diameter) is pressed into the material while load and displacement are recorded [99]. | Elastic modulus and hardness calculated from the load-displacement curve [99]. | Soft, hydrated biomaterials (hydrogels, cartilage), and stiff, mineralized tissues (bone) [99]. |
| Macroscale Testing (e.g., Unconfined Compression) | A cylindrical sample is compressed between two plates, and the force required for deformation is measured [99]. | Stress-strain relationship used to determine the compressive elastic modulus [99]. | Homogeneous, engineered materials and soft tissues; requires samples of sufficient size [99]. |
The comparative study confirmed that Pipette Aspiration and Nanoindentation measure similar elastic moduli for silicones and hydrogels in the range of 10 to 400 kPa, and these results are consistent with moduli from bulk testing [99]. This validation is critical for building confidence in in vitro mechanical data, though it must still be followed by in vivo performance assessment.
Section 3: The Scientist's Toolkit: Essential Research Reagent Solutions
The following table catalogs key materials and their functions essential for conducting rigorous in vitro and in vivo biomaterial research:
| Research Reagent / Material | Function in Experimentation |
|---|---|
| Cross-linked Porcine Small Intestine (CLPSI) | A biomaterial scaffold used for orthopedic soft tissue augmentation; demonstrated favorable in vitro cell viability and in vivo clinical application potential [98]. |
| Non-cross-linked Porcine Small Intestine (NCLPSI) | A biomaterial scaffold showing superior cell infiltration in vitro and the most rapid replacement with regenerative tissue in vivo in rat models [98]. |
| Silicone Elastomers (Ecoflex, Dragon Skin) | Model materials with tunable stiffness (20-400 kPa) used for method validation and as tissue analogs in mechanical testing studies [99]. |
| Polyacrylamide Hydrogels | Synthetic hydrogels with moduli from 10-50 kPa; used in cell culture studies to investigate substrate stiffness effects and as tissue analogs [99]. |
| MTT / XTT Assay Kits | Colorimetric assays that measure the reduction of tetrazolium salts by metabolically active cells, serving as a key indicator of cell viability in cytotoxicity testing [21]. |
| Cell Culture Media & Extraction Solvents | Physiological saline, vegetable oil, and cell culture medium are used to prepare extracts of medical devices to evaluate leachable chemicals [21]. |
Section 4: The Integrated Workflow: From Bench to Bedside
The most successful translational research strategies employ a sequential, integrated workflow that leverages the strengths of both in vitro and in vivo approaches. This pathway systematically de-risks the development process.
Diagram 2: The Integrated In Vitro / In Vivo Research Workflow
This non-negotiable integrated approach is exemplified in the drug development pipeline. Researchers start with in vitro tests on cells in lab dishes to identify promising drug candidates and evaluate basic safety [2] [38]. Successful candidates then proceed to in vivo animal studies to understand effects in a whole-organism context, including complex interactions like immune response and drug metabolism [2] [45]. Finally, human clinical trials, which are the ultimate form of in vivo testing, are conducted. These often use randomized controlled trials (RCTs), where participants are randomly assigned to treatment or control groups, and blinding is used to eliminate bias [2]. This stepwise process is vital: approximately 30% of drug candidates fail in human trials due to safety problems, and another 60% fail due to lack of efficacy, underscoring the predictive limitations of isolated systems [2].
The dichotomy between in vitro and in vivo methodologies presents a false choice. In vitro models provide unmatched control, speed, and cost-efficiency for foundational discovery and mechanistic studies, while in vivo systems offer the indispensable complexity of whole-organism physiology. As the data shows, biomaterials that excel in Petri dishes can fail in living organisms, and drugs that seem promising on cell lines may be ineffective or toxic in patients. Therefore, a combined approach is not merely an academic ideal but a practical necessity. It is the only path that fully respects the ethical imperative of the 3Rs (Replacement, Reduction, and Refinement) in animal testing while rigorously validating safety and efficacy. For researchers and drug development professionals committed to delivering safe and effective medical solutions, this integrated pathway remains the non-negotiable gold standard.
In the realm of biomedical research and drug development, in vitro and in vivo testing represent two fundamental paradigms for evaluating the safety and efficacy of new compounds, drugs, and biomaterials. These complementary approaches provide distinct insights at different stages of the preclinical development pipeline. In vitro (Latin for "in the glass") refers to experiments conducted with microorganisms, cells, or biological molecules outside their normal biological context, in controlled environments such as petri dishes or test tubes [63]. These studies are characterized by their focus on isolated components of living organisms, allowing researchers to investigate specific biological mechanisms without the complexity of a whole living system.
In contrast, in vivo (Latin for "within the living") testing involves experiments performed on whole, living organisms, typically animals such as rodents, zebrafish, or non-human primates [63]. These studies provide a comprehensive view of how an experimental substance behaves in a complex physiological environment, where multiple biological systems interact simultaneously. The fundamental distinction between these approaches lies in their biological complexity: in vitro systems offer precision and control for mechanistic studies, while in vivo systems provide physiological relevance by accounting for the intricate interactions that occur within a complete living organism. This comparison guide will objectively examine the key parameters, applications, and methodological considerations for both testing approaches within the context of biomaterial testing results comparison research.
Comparative Analysis: Applications and Methodologies
The selection between in vitro and in vivo testing methodologies depends heavily on the research objectives, regulatory requirements, and stage of development. In pharmaceutical applications, in vitro studies typically serve as the first line of investigation, providing preliminary data on compound efficacy and toxicity before progressing to more complex and costly in vivo models [63]. These studies are particularly valuable for high-throughput screening, where thousands of compounds can be rapidly tested against specific molecular targets or cellular pathways. The pharmaceutical industry represents the largest end-user segment for in vitro toxicity testing, driven by demands for early-stage toxicity screening in drug development and regulatory pressures to reduce animal testing [100].
In vivo testing remains indispensable for assessing complex physiological responses that cannot be adequately modeled in isolated systems. Certain toxicological endpoints—including carcinogenicity, developmental and reproductive toxicity (DART), immunotoxicity, and systemic toxicity—are still primarily evaluated through in vivo studies due to the current limitations of alternative methods [101]. For instance, chronic toxicity tests, which are carried out with a minimum of one rodent and one non-rodent species where the test compound is administered for more than 90 days, provide crucial information about the long-term effects of substances that cannot be fully replicated in current in vitro systems [101]. The in vivo toxicology market continues to grow, with an estimated value of $5.0 billion in 2020 projected to reach $6.6 billion by 2025, reflecting its ongoing importance in safety assessment [101].
Table 1: Fundamental Characteristics and Applications of In Vitro vs. In Vivo Testing
| Parameter | In Vitro Testing | In Vivo Testing |
|---|---|---|
| Definition | Experiments conducted on cells, tissues, or biological molecules outside their normal biological context [63] | Experiments performed on whole, living organisms [63] |
| Primary Applications | - Early-stage drug screening- Cellular and molecular studies- High-throughput toxicity screening- Mechanism of action studies | - Complex toxicity assessments (carcinogenicity, DART)- Systemic effects evaluation- Pharmacokinetics and pharmacodynamics- Efficacy validation |
| Typical Models | - Cell cultures (2D and 3D)- Organ-on-chip systems- Tissue explants- Bacterial cultures | - Rodents (mice, rats)- Zebrafish- Non-human primates- Other mammalian species |
| Regulatory Status | Increasingly accepted for specific endpoints; OECD guidelines available for various assays [100] | Remains mandatory for many regulatory submissions; required for complex endpoints [101] |
| Market Position | $10.04 billion market in 2025 (13.5% CAGR projected) [100] | $5.0 billion market in 2020 (11.5% CAGR projected) [101] |
Key Parameter Comparison Tables
Technical and Experimental Parameters
The methodological differences between in vitro and in vivo approaches significantly influence experimental design, data interpretation, and translational potential. In vitro systems excel in experimental control and reproducibility, as researchers can precisely manipulate environmental conditions such as temperature, pH, nutrient availability, and compound concentration [102] [103]. This level of control enables detailed mechanistic studies but comes at the cost of reduced physiological relevance, as these systems cannot fully replicate the complex cellular interactions, metabolic processes, and systemic responses that occur in living organisms [63].
In vivo testing provides comprehensive physiological context but introduces substantial biological variability that must be accounted for in experimental design and statistical analysis. The presence of intact biological barriers (e.g., gastrointestinal tract, blood-brain barrier), functional immune systems, and organ-organ interactions in living organisms creates a more accurate representation of human physiological responses but complicates the isolation of specific mechanistic pathways [26]. Recent advances in statistical frameworks like SynergyLMM have improved the analysis of in vivo combination experiments by accounting for longitudinal data structure and inter-animal heterogeneity, thereby enhancing the robustness of conclusions drawn from these complex studies [26].
Table 2: Technical and Methodological Comparison
| Technical Parameter | In Vitro Testing | In Vivo Testing |
|---|---|---|
| Environmental Control | High precision control over temperature, pH, medium composition [102] | Limited control; subject to organism's homeostatic regulation |
| Biological Complexity | Reduced complexity; focuses on isolated components [63] | Full physiological complexity with interacting systems [63] |
| Experimental Duration | Typically shorter (hours to days) [102] [103] | Typically longer (days to months) [101] |
| Data Reproducibility | Generally higher due to controlled conditions [63] | Lower due to biological variability between subjects [26] |
| Throughput Capacity | High-throughput screening possible [100] | Lower throughput due to cost and ethical considerations |
| Biological Relevance | Limited to specific pathways or mechanisms [63] | High; captures systemic responses and complex interactions [63] |
| Statistical Considerations | Less complex statistical models required | Advanced models needed (e.g., mixed effects models) to account for variability [26] |
Operational and Regulatory Parameters
From an operational perspective, in vitro and in vivo testing differ significantly in cost structure, timeline, ethical considerations, and regulatory requirements. In vitro methods generally offer lower costs and faster results, making them particularly suitable for early-stage screening where large numbers of compounds must be evaluated rapidly [63] [100]. The in vitro toxicity testing market is experiencing robust growth (13.5% CAGR), driven by technological advancements in cell-based assays, organ-on-chip systems, and increasing regulatory pressure to reduce animal testing [100]. These methods align with the 3Rs principles (Replacement, Reduction, and Refinement) in animal research and are increasingly accepted by regulatory agencies for specific endpoints.
In vivo testing involves higher costs and longer timelines but remains necessary for comprehensive safety assessment and regulatory approval of many products. The in vivo toxicology market continues to grow at 11.5% CAGR, reflecting its ongoing importance despite the development of alternative methods [101]. Certain tests—including those for carcinogenicity, developmental and reproductive toxicity (DART), and immunotoxicity—are still primarily conducted in vivo due to the current limitations of non-animal methods [101]. However, regulatory frameworks such as EU REACH are increasingly encouraging the use of alternative methods, including in silico approaches like the CATMoS model for acute oral toxicity prediction, which represents a shift toward integrated testing strategies that leverage multiple approaches [104].
Table 3: Operational and Regulatory Comparison
| Operational Parameter | In Vitro Testing | In Vivo Testing |
|---|---|---|
| Cost Factors | Lower cost per sample; minimal consumable expenses | High costs associated with animal purchase, housing, and care |
| Time Requirements | Rapid results (typically hours to days) [63] | Extended study durations (days to months) [101] |
| Ethical Considerations | Minimal ethical concerns; no animal use [63] [100] | Significant ethical considerations; strict oversight required |
| Regulatory Acceptance | Growing acceptance for specific endpoints [100] | Well-established and often mandatory for many applications [101] |
| Standardization Level | Increasing standardization through OECD guidelines [100] | Highly standardized protocols for specific test types [101] |
| Personnel Requirements | Standard laboratory technical skills | Specialized training in animal handling and procedures |
Experimental Protocols and Workflows
In Vitro Release Testing Protocol for Liposomal Formulations
The assessment of drug release characteristics from nanomedicine formulations like liposomes represents a sophisticated application of in vitro methodology. This protocol, adapted from recent research on making release data AI-ready, outlines a standardized approach for evaluating in vitro release (IVR) from liposomal formulations [102] [103].
Materials and Reagents:
- Liposome formulation with known critical quality attributes (CQAs: particle size, zeta potential, drug loading)
- Appropriate release media (e.g., phosphate-buffered saline, simulated biological fluids)
- Dialysis membranes or diffusion cells with appropriate molecular weight cut-off
- Temperature-controlled shaking water bath or orbital shaker
- Sampling apparatus (autosampler or manual sampling equipment)
- Analytical instrument for drug quantification (e.g., HPLC, UV-Vis spectrophotometer)
Methodology:
- Formulation Characterization: Determine critical quality attributes of the liposome formulation, including particle size, polydispersity index, zeta potential, and drug loading prior to release testing [102] [103].
- Release Media Selection: Choose appropriate release media based on the intended administration route and site of action. Consider factors such as pH, osmolarity, and sink conditions [102].
- Apparatus Setup: Assemble the release apparatus (dialysis system, flow-through cell, or paddle apparatus) and equilibrate to the appropriate temperature (typically 37°C for systemic administration) [102] [103].
- Sample Introduction: Introduce a precise volume of liposome formulation into the release apparatus containing the pre-equilibrated media.
- Sampling Time Points: Collect samples at predetermined time intervals (e.g., 0.5, 1, 2, 4, 8, 12, 24 hours) while maintaining sink conditions throughout the experiment.
- Sample Analysis: Process samples as needed (e.g., filtration, dilution) and analyze using appropriate analytical methods to quantify drug concentration.
- Data Processing: Calculate cumulative drug release (%) versus time and plot release profiles. Fit appropriate mathematical models (e.g., Weibull, Higuchi, zero-order) to characterize release kinetics [103].
Quality Control Considerations:
- Validate analytical methods for accuracy, precision, and specificity
- Include appropriate controls (e.g., free drug solution) to assess method performance
- Maintain consistent hydrodynamic conditions throughout the experiment
- Document all critical process parameters and material attributes for data traceability [102]
This standardized protocol generates reproducible release data that can be used for quality assessment, formulation optimization, and with the advent of machine learning approaches, predictive modeling of formulation performance [102] [103].
In Vivo Drug Combination Efficacy Protocol
The evaluation of drug combination effects in oncology represents a complex in vivo application that requires careful experimental design and statistical analysis. This protocol, adapted from the SynergyLMM framework, outlines a comprehensive approach for assessing combination therapy efficacy in tumor-bearing mouse models [26].
Materials and Animals:
- Immunocompromised mice (e.g., nude, NSG) or immunocompetent syngeneic models
- Cancer cell lines or patient-derived xenograft (PDX) models
- Test compounds (single agents and combinations)
- Caliper for tumor measurement or in vivo imaging system
- Appropriate vehicle controls
Methodology:
- Tumor Implantation: Implant cancer cells or tumor fragments subcutaneously into appropriate mouse models and allow tumors to establish to a predetermined volume (typically 100-200 mm³).
- Randomization: Randomly assign animals to treatment groups (control, single agents, combination) ensuring similar initial tumor size distributions across groups.
- Dosing Regimen: Administer treatments according to predetermined schedules and routes (oral, intraperitoneal, intravenous) at the maximum tolerated doses or clinically relevant exposures.
- Tumor Measurement: Measure tumor dimensions regularly (2-3 times weekly) using calipers or via in vivo imaging for luciferase-expressing models. Record body weights simultaneously to monitor toxicity.
- Endpoint Determination: Continue treatment until predetermined endpoint (e.g., maximum tumor volume, study duration) is reached.
- Data Collection: Record longitudinal tumor volume data and any observations of toxicity or morbidity.
Statistical Analysis Using SynergyLMM Framework:
- Data Preprocessing: Normalize tumor volumes to baseline measurements at treatment initiation.
- Model Selection: Choose appropriate growth kinetic model (exponential or Gompertz) based on tumor growth characteristics.
- Mixed Effects Modeling: Fit (non-)linear mixed effects models to account for inter-animal variability while estimating group-level treatment effects [26].
- Synergy Assessment: Calculate synergy scores (SS) using appropriate reference models (Bliss independence, Highest Single Agent, or Response Additivity) with statistical significance testing [26].
- Time-Resolved Analysis: Evaluate synergy patterns across different time points to identify dynamic changes in combination effects [26].
- Model Diagnostics: Perform diagnostic checks to verify model assumptions and identify potential outliers or influential observations [26].
This comprehensive approach enables robust evaluation of combination therapy effects while accounting for the longitudinal nature of tumor growth data and inter-animal heterogeneity, providing higher confidence in the identification of truly synergistic combinations [26].
Visualization of Testing Workflows
In Vitro Release Testing Workflow
The following diagram illustrates the key steps and decision points in a standardized in vitro release testing workflow for liposomal formulations, highlighting the integration of machine learning approaches for data analysis:
In Vitro Release Testing Workflow
In Vivo Combination Therapy Assessment
The following diagram outlines the comprehensive statistical framework for analyzing in vivo drug combination experiments, highlighting the longitudinal data analysis and synergy assessment components:
In Vivo Combination Therapy Assessment
Essential Research Reagent Solutions
The following table details key reagents, models, and tools essential for conducting robust in vitro and in vivo studies in the context of biomaterial and drug formulation testing:
Table 4: Essential Research Reagents and Tools
| Reagent/Tool | Application | Function | Examples/Specifications |
|---|---|---|---|
| Liposome Formulations | In vitro release testing | Model drug delivery system for evaluating release kinetics [102] [103] | Varied lipid compositions, cholesterol content, surface modifications |
| Cell-Based Assays | In vitro toxicity screening | Assessment of cellular viability, proliferation, and toxicity endpoints [100] | High-throughput screening formats, stem cell-based assays (e.g., ToxTracker) |
| Organ-on-Chip Systems | Advanced in vitro modeling | Recreation of human organ physiology for predictive toxicity testing [100] | Multi-organ microphysiological systems, PhysioMimix platforms |
| Rodent Models | In vivo toxicology and efficacy | Comprehensive safety and efficacy assessment in mammalian systems [101] | Mice, rats; genetically engineered models, patient-derived xenografts |
| Zebrafish Models | Alternative in vivo testing | Bridge between in vitro and mammalian models; developmental studies [101] [63] | Embryos (<5-6 dpf) not considered experimental animals in EU Directive |
| Machine Learning Tools | Data analysis and prediction | Pattern recognition in complex datasets; prediction of release profiles and toxicity [102] [103] [104] | CATMoS for acute toxicity prediction, SynergyLMM for combination studies |
| Analytical Instruments | Drug quantification and characterization | Quantification of drug release, formulation characterization [102] | HPLC systems, UV-Vis spectrophotometers, dynamic light scattering |
The comparative analysis of in vitro and in vivo testing approaches reveals a complementary relationship rather than a competitive one in biomedical research and drug development. In vitro systems provide unparalleled control, reproducibility, and throughput for early-stage screening and mechanistic studies, while in vivo models offer irreplaceable physiological context for evaluating complex systemic responses and validating efficacy and safety. The ongoing advancement of both approaches—including the development of sophisticated in vitro systems like organ-on-chip technologies and the refinement of in vivo study designs through advanced statistical frameworks—continues to enhance their respective values in the research continuum.
The future of preclinical testing lies not in choosing between these approaches but in strategically integrating them through fit-for-purpose application and leveraging emerging technologies such as machine learning and artificial intelligence to bridge the gaps between different data types [102] [103] [26]. Furthermore, the growing regulatory acceptance of alternative methods and the development of integrated testing strategies promise to optimize the use of both in vitro and in vivo approaches, maximizing scientific insight while adhering to ethical principles and efficient resource utilization [104] [100]. As both technologies evolve, this complementary relationship will continue to drive innovations in drug development and safety assessment, ultimately accelerating the delivery of new therapies to patients.
The development of new medical devices and implants is a complex, costly, and time-intensive process that relies heavily on accurate preclinical assessment [105]. Biomaterials, defined as "nonviable materials used in a medical device, intended to interact with biological systems," must undergo rigorous testing to ensure safety and efficacy before clinical use [105] [106]. The traditional approach involves extensive in vitro testing followed by validation in animal models, creating a significant bottleneck in biomaterial development [7]. This process is further complicated by the fact that complex host responses to biomaterials involve multiple interacting cell types and dynamic inflammatory processes that are difficult to model accurately in laboratory settings [7].
Establishing a reliable ranking system for biomaterials based on their predicted in vivo performance requires sophisticated assay development and computational analysis methods [7]. This comparison guide examines current methodologies for correlating in vitro findings with in vivo outcomes, providing researchers with validated approaches for predicting biomaterial performance while reducing reliance on animal testing. By implementing these strategies, the biomaterials community can accelerate the development of safer, more effective medical devices while addressing the growing ethical and economic imperatives to reduce animal experimentation [107].
Biomaterial-Tissue Interaction Fundamentals
The host response to implanted biomaterials is a critical determinant of their clinical success. This response varies significantly based on material composition, surface characteristics, and structural properties [7]. Understanding these fundamental interactions provides the foundation for developing predictive in vitro models.
Generations of Biomaterials
Biomaterials have evolved through three distinct generations, each with different tissue interaction profiles:
- First-generation (bioinert materials): Provide mechanical support while minimizing interaction with host tissues (e.g., titanium, steel) [105]
- Second-generation (bioactive and bioresorbable): Designed to interact with host tissues through interfacial bonding (e.g., bioactive ceramics, bioglasses) [105]
- Third-generation (bioactive and bioresorbable): Actively stimulate cellular responses for tissue repair and regeneration through interconnected porous architecture [105]
Key Cellular Players in Host Response
Macrophages are central regulators of the host response to biomaterials, secreting biochemical signals that directly influence tissue remodeling and producing proteases that actively remodel degradable biomaterials [7]. The polarization state of macrophages (classically activated M1 or alternatively activated M2 phenotypes) significantly influences the remodeling outcome, with M2 macrophages associated with constructive remodeling and M1 macrophages linked to foreign body reactions [7].
Table: Key Cell Types in Biomaterial-Host Interactions
| Cell Type | Role in Host Response | Impact on Remodeling Outcome |
|---|---|---|
| Macrophages | Secretion of cytokines, chemokines, and proteases; formation of foreign body giant cells | Central regulators: M2 phenotype promotes constructive remodeling; M1 promotes fibrosis [7] |
| Fibroblasts | Deposition and organization of connective tissue; formation of fibrotic capsule | Excessive activity leads to encapsulation and isolation of implant [7] |
| Lymphocytes | Modulation of immune response through cytokine signaling | Can amplify inflammatory responses to biomaterials [7] |
| Osteoblasts (bone applications) | Bone matrix formation and mineralization | Critical for osseointegration of orthopedic and dental implants [105] |
Established Methodologies for In Vitro to In Vivo Correlation
Macrophage Response Profiling with In Silico Analysis
A pioneering approach developed by researchers combines in vitro macrophage response characterization with quasi-mechanistic in silico analysis to predict in vivo outcomes [7]. This method addresses the limitation of traditional in vitro assays that often fail to capture the complexity of in vivo responses.
Experimental Protocol:
- Human monocyte-derived macrophages are cultured with test biomaterials for specified periods
- Dynamic inflammatory response is characterized through multiplexed cytokine/chemokine profiling
- In silico analysis using Principal Component Analysis (PCA) and Dynamic Network Analysis (DyNA) identifies response patterns
- Pattern matching associates in vitro signatures with known in vivo outcomes from reference materials
- Prediction model is validated against independent test materials with known in vivo performance [7]
Key Application: This approach successfully distinguished between materials that would elicit constructive remodeling versus those that would trigger a foreign body reaction in rodent models, with in vitro macrophage responses accurately predicting the dominant macrophage polarization (M1/M2) observed in vivo [7].
Standardized Cytotoxicity Testing According to ISO 10993-5
For initial biocompatibility screening, standardized cytotoxicity testing provides a foundational assessment of biomaterial safety [30]. The ISO 10993-5 standard outlines three primary test formats: extract, direct contact, and indirect contact tests [30].
Detailed MTT Assay Protocol:
- Extract Preparation: Biomaterial samples are incubated in Dulbecco's Modified Eagle Medium (DMEM) supplemented with fetal bovine serum (FBS) using surface area-to-volume ratios specified in ISO standards
- Cell Culture: L-929 mouse fibroblast cells (or other mammalian cell lines) are cultured in standardized conditions (37°C, 5% CO₂) for 24-72 hours
- Exposure: Cell monolayers are exposed to biomaterial extracts for specified periods (typically 24 hours)
- Viability Assessment: MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) is added to cultures and incubated for 2-4 hours
- Formazan Solubilization: Insoluble purple formazan crystals are dissolved using organic solvents (isopropanol or DMSO)
- Quantification: Absorbance is measured at 570nm, with reference wavelength ~650nm
- Calculation: Cell viability percentage is calculated relative to negative controls [30]
Interpretation Criteria: Cell viability >70% relative to controls is generally considered non-cytotoxic, with viability <70% indicating potential cytotoxicity requiring further investigation [30].
Table: Cytotoxicity Assessment of Magnesium-Based Biomaterials
| Biomaterial | Extract Concentration | Cell Viability (%) | Cytotoxicity Classification | Key Findings |
|---|---|---|---|---|
| Mg-1%Sn-2%HA composite | 100% | 71.51% | Non-cytotoxic | Minimal morphological changes observed [30] |
| Mg-1%Sn-2%HA composite | 50% | 84.93% | Non-cytotoxic | Dose-response relationship evident [30] |
| Mg-1%Sn-2%HA composite | 25% | 93.20% | Non-cytotoxic | Near-complete viability restoration [30] |
| Mg-1%Sn-2%HA composite | 12.5% | 96.52% | Non-cytotoxic | Confirmed safety at diluted concentrations [30] |
The Chick Chorioallantoic Membrane (CAM) Model as a 3R-Compliant Intermediate
The CAM model represents a promising intermediate between in vitro and in vivo testing that aligns with the 3R principles (Replacement, Reduction, and Refinement of animal experiments) [107].
Validation Study Protocol:
- Egg Preparation: Fertilized specific pathogen-free chicken eggs are incubated at 37°C and 70-80% humidity
- Window Preparation: At embryonic development day (EDD) 8, a window is introduced in the eggshell and the shell membrane is carefully removed
- Biomaterial Implantation: At EDD 9, test biomaterials (approximately 40mg) are transplanted onto the CAM
- Harvest: After 24 hours (EDD 10), the CAM tissue with implanted biomaterial is harvested and fixed in 4% buffered formaldehyde
- Histopathological Evaluation: Tissue sections are stained with hematoxylin and eosin and scored according to DIN EN ISO 10993-6 criteria [107]
Direct Validation: This model has been rigorously compared with subcutaneous implantation in rats, demonstrating complete comparability in tissue responses with no statistical differences in histopathological scoring [107]. This suggests that bioincompatible biomaterials can be effectively identified using the CAM model before proceeding to more complex mammalian systems.
Advanced Biomaterial Assessment Technologies
Artificial Intelligence in Biomaterial Development
AI approaches are transforming biomaterial research by enabling property prediction and structure generation beyond conventional trial-and-error methods [105]. Machine learning (ML) applications in material science generally fall into two categories:
- Property Prediction (Forward Problem Design): Forecasting desired material properties by evaluating candidate materials from extensive libraries [105]
- Structure Generation (Inverse Problem Design): Designing optimal biomaterial structures to achieve specific target properties [105]
These approaches significantly accelerate the material design process and facilitate exploration of novel combinations beyond existing knowledge boundaries [105].
3D Printing for Complex Tissue Engineering Scaffolds
Advanced manufacturing technologies, particularly 3D printing, enable precise control over scaffold architecture for neural tissue engineering applications [53]. Different biomaterials offer distinct advantages for specific applications:
Table: Comparison of 3D-Printable Biomaterials for Neural Tissue Engineering
| Biomaterial | Type | Key Properties | Printability | Cell Viability | Best Applications |
|---|---|---|---|---|---|
| Polylactic Acid (PLA) | Synthetic polymer (thermoplastic) | Excellent thermal stability, tunable degradability [53] | High resolution and shape fidelity [53] | Moderate | Structural nerve guides, support scaffolds |
| Polycaprolactone (PCL) | Synthetic polymer (thermoplastic) | Mechanical elasticity, long-term degradation, low melting temperature [53] | Good printing stability [53] | Moderate | Long-term implantable nerve conduits |
| Gelatin Methacrylate (GelMA) | Natural-derived hydrogel | Biofunctional motifs, tunable mechanical properties, biomimetic [53] | Lower shape fidelity requires support [53] | High (superior after 7 days) [53] | Cell-laden constructs, bioactive interfaces |
| Filaflex (FF) | Conductive thermoplastic polyurethane | High flexibility, electroconductive properties [53] | Good flexibility retention [53] | Under investigation | Neural interfaces requiring conductivity |
| Flexdym (FD) | Styrene-ethylene/butylene-styrene (SEBS) | Flexible, stretchable, biocompatible [53] | Excellent viscoelastic properties [53] | Under investigation | Soft tissue interfaces, compliant implants |
Corrosion Modeling for Metallic Implants
For biodegradable metallic implants, particularly magnesium-based alloys, predictive corrosion modeling is essential for determining in vivo performance [108]. Phenomenological corrosion models incorporate factors such as:
- Strain-assisted corrosion mechanisms
- In vivo environmental factors
- Material composition effects
- Mechanical loading conditions
These models are calibrated using in vitro data and validated through in vivo implantation studies, creating a predictive framework for implant degradation behavior [108].
Integrated Workflow for Biomaterial Ranking
The following diagram illustrates a comprehensive workflow for establishing biomaterial rankings through integrated in vitro and in vivo validation:
Workflow for Biomaterial Ranking Validation
The Scientist's Toolkit: Essential Research Reagents and Materials
Table: Key Research Reagent Solutions for Biomaterial Testing
| Reagent/Material | Function/Application | Specific Examples | Experimental Considerations |
|---|---|---|---|
| L-929 Mouse Fibroblast Cells | Standardized cell line for cytotoxicity testing (ISO 10993-5) [30] | ATCC CCL-1 | Sensitive indicator of cytotoxic effects; well-characterized response |
| Human Monocyte-Derived Macrophages | Predictive model of host immune response to biomaterials [7] | Primary cells from peripheral blood mononuclear cells (PBMCs) | Donor variability requires multiple replicates; polarization state critical |
| Dulbecco's Modified Eagle Medium (DMEM) | Extraction medium for cytotoxicity testing [30] | With fetal bovine serum (FBS) supplementation | Serum content affects extraction efficiency; standardized ratios required |
| MTT Reagent | (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) for cell viability assessment [30] | Mitochondrial dehydrogenase activity measurement | Requires solubilization step; light-sensitive; crystal formation indicates viability |
| Specific Pathogen-Free (SPF) Chicken Eggs | CAM model for intermediate in vivo testing [107] | Lohmann strains | Embryonic day critical for implantation success; windowing technique sensitive |
| Ti Grade 2 Powder | Metamaterial fabrication for auxetic implants [109] | Selective laser melting processing | Lower elastic modulus vs. Ti6Al4V; better mechanical compatibility with bone |
| GelMA Hydrogel | Bioink for 3D printed neural scaffolds [53] | Photocrosslinkable with LAP initiator | Concentration affects mechanical properties; degree of methacrylation critical |
The establishment of reliable biomaterial ranking systems requires integrated approaches that correlate standardized in vitro assessments with in vivo outcomes. The methodologies presented in this guide - from macrophage response profiling with in silico analysis to the validated CAM model - provide researchers with practical frameworks for predicting biomaterial performance while adhering to 3R principles.
As the field advances, the integration of artificial intelligence with high-throughput experimental data promises to further enhance our predictive capabilities [105]. This will enable more efficient screening of novel biomaterial combinations and accelerate the development of next-generation medical devices that demonstrate improved clinical outcomes. By implementing these validated correlation strategies, researchers can establish robust biomaterial ranking systems that effectively bridge the gap between laboratory findings and clinical performance.
The journey from a laboratory concept to a clinically approved therapy relies on a critical pipeline of preclinical testing. Within this pipeline, in vitro (conducted in controlled laboratory environments outside a living organism) and in vivo (conducted within a living organism) models represent two foundational approaches. The central challenge for researchers and drug development professionals lies in strategically balancing these methods. This balance must carefully weigh the predictive power necessary for clinical success against significant ethical considerations and substantial resource constraints, including cost, time, and specialized expertise. A thorough cost-benefit analysis is not merely an academic exercise but a practical necessity for the efficient and responsible development of new medical interventions [110] [111].
Defining the Paradigms: In Vitro vs. In Vivo Testing
At its core, the distinction between these two approaches is defined by the experimental environment.
- In Vitro Testing: The term is Latin for "in glass," describing experiments performed on isolated cells, tissues, or biological molecules in an artificial, controlled setting, such as a petri dish or test tube [2] [38]. This methodology allows scientists to examine specific biological phenomena in isolation, free from the complex interactions of a whole organism.
- In Vivo Testing: Meaning "within the living" in Latin, this involves experiments carried out in or on a whole living organism, such as laboratory mice, rats, or humans in clinical trials [2] [112]. It provides a holistic view of how a biological system responds within a natural, physiologically complex environment.
The following visual workflow outlines the typical role of each method in the sequential and iterative process of therapeutic development:
Quantitative Cost-Benefit Analysis
A strategic balance between in vitro and in vivo models is crucial because over-reliance on either has led to costly translational failures. Studies indicate that approximately 30% of drug candidates fail in human clinical trials due to adverse side effects not predicted in earlier tests, and a further 60% fail due to a lack of the desired efficacy [2]. This high failure rate underscores the limitations of existing models and the financial implications of an imperfect testing strategy.
Comparative Analysis of Testing Models
The table below summarizes the core advantages, limitations, and associated costs of each approach, providing a framework for a direct comparison.
| Aspect | In Vitro Testing | In Vivo Testing |
|---|---|---|
| Primary Advantages | Controlled, simplified environment; High throughput; Cost-effective for early screening; Avoids ethical concerns of animal use [110] [112]. | High physiological relevance; Captures complex system interactions; Reveals unexpected side effects; Provides long-term effect data [110] [38]. |
| Key Limitations | Poor replication of whole-organism complexity; Lacks biokinetics (ADME); High false-positive/negative rates; Uses often abnormal (e.g., cancerous) cell lines [110] [23]. | Extremely high cost and time-consuming; Significant ethical concerns and regulations; Physiological differences between animals and humans; Requires advanced personnel training [110] [111]. |
| Relative Cost | Low to Moderate | Very High |
| Experimental Duration | Short (days to weeks) | Long (months to years) |
| Predictive Power | Low to Moderate (mechanistic insights) | Moderate to High (but not perfect) |
The High Cost of In Vivo Research
The resource intensity of in vivo studies is not merely theoretical. A systematic review of a specific medical application—synthetic or biosynthetic nerve scaffolds for peripheral nerve repair—quantified the immense investment in preclinical animal studies. The review analyzed 416 studies that used eight different animal species and over 65 evaluation methods. The total estimated cost for these preclinical studies alone reached $61,264,910 [111]. This figure highlights the massive financial burden associated with in vivo testing long before a product ever reaches human trials.
Establishing Predictive Power: The Role of IVIVC
To bridge the gap between laboratory models and human outcomes, scientists use In Vitro-In Vivo Correlation (IVIVC). The U.S. Food and Drug Administration (FDA) defines IVIVC as "a predictive mathematical model describing the relationship between an in vitro property of a dosage form (typically the rate or extent of drug dissolution) and a relevant in vivo response (typically plasma drug concentration or amount absorbed)" [113] [114]. A robust IVIVC is a powerful tool that can reduce the need for certain clinical studies and accelerate development.
Levels and Regulatory Value of IVIVC
IVIVC models are categorized by their predictive strength, which determines their regulatory utility.
| Level | Definition | Predictive Value & Regulatory Acceptance |
|---|---|---|
| Level A | A point-to-point correlation between in vitro dissolution and the in vivo absorption profile [113] [114]. | High. Most preferred by regulators; can support biowaivers for formulation changes [114]. |
| Level B | A statistical comparison using mean in vitro dissolution time and mean in vivo residence or absorption time [113] [114]. | Moderate. Less robust; usually requires additional in vivo data [114]. |
| Level C | A single point correlation (e.g., relating a dissolution time point to a pharmacokinetic parameter like Cmax or AUC) [113] [114]. | Low. Least rigorous; insufficient for biowaivers but useful for early development insights [114]. |
Challenges in Correlation for Complex Formulations
Establishing a reliable IVIVC is particularly challenging for complex drug delivery systems, such as Lipid-Based Formulations (LBFs), which are designed to improve the absorption of poorly water-soluble drugs. The dynamic processes of lipid digestion, micelle formation, and permeation are not fully captured by traditional in vitro dissolution tests [113]. Case studies on drugs like fenofibrate and cinnarizine have shown repeated failures in establishing predictive IVIVCs, with in vitro models unable to distinguish between the in vivo performance of different LBFs or accurately predict absorption patterns [113]. This underscores the limitations of current in vitro models and the consequent reliance on costly in vivo studies for these formulations.
Ethical Considerations in Testing Choices
Ethical constraints form a critical dimension of the cost-benefit analysis, heavily influencing the choice of testing model.
The "3Rs" Principle and Animal Welfare
The use of animals in research is governed by the "3Rs" principle: Replace, Reduce, and Refine [112]. This ethical framework mandates that researchers should seek to replace animal models with alternatives (like advanced in vitro models) where possible, reduce the number of animals used to a minimum, and refine procedures to minimize animal suffering [112]. These principles are not only ethical imperatives but also help manage the high costs and regulatory scrutiny associated with animal studies.
The Status of Human Embryos and Embryo-Like Structures
Research on human embryos presents another profound ethical challenge. Many countries operate under a "gradualist" view, where the moral status of the embryo increases with its biological development [115]. This has led to regulations like the "14-day rule," a international standard prohibiting the culturing of human embryos for research beyond 14 days of development [115]. With advancing technology, there is now debate about extending this limit to 28 days to study crucial stages of organ development, while also acknowledging that a less controversial alternative (research on aborted tissues) becomes available after 28 days [115].
Furthermore, the emergence of embryo-like structures (ELSs) created from stem cells blurs ethical lines. Current consensus suggests that integrated ELSs (with the potential to form a full organism) should be subject to similar rules as natural embryos if they pass certain tests, while non-integrated ELSs may warrant a lower level of moral concern [115]. This evolving landscape requires continuous ethical scrutiny.
The Researcher's Toolkit: Essential Materials and Reagents
The execution of both in vitro and in vivo experiments relies on a suite of specialized tools and reagents. The following table details key materials essential for biomaterial testing and basic research in this field.
| Research Reagent / Material | Primary Function in Testing |
|---|---|
| Polymer & Natural Biomaterials | Used to create synthetic or biosynthetic scaffolds (e.g., for nerve repair). Testing equipment evaluates their degradation rates, mechanical strength, and biointegration [111] [116]. |
| Metallic Biomaterials | Used for implants (e.g., joint replacements, stents). Testing equipment assesses properties like tensile strength, fatigue resistance, and corrosion resistance [116]. |
| Ceramic Biomaterials | Common in dental and orthopedic applications. Testing focuses on properties like wear resistance, bioactivity, and compressive strength [116]. |
| USP Dissolution Apparatus | A standard in vitro tool to measure the rate and extent of drug release from a dosage form under standardized conditions, forming the basis for IVIVC [113]. |
| pH-Stat Lipolysis Assay | An advanced in vitro tool that simulates the digestion of lipid-based formulations in the gastrointestinal tract, crucial for predicting the performance of LBFs [113]. |
Decision Framework and Future Perspectives
Choosing between in vitro and in vivo models is not a binary decision but a strategic one. The following diagram outlines a logical framework to guide this choice based on project stage, key constraints, and objectives.
The future of biomaterial testing lies in the development of more sophisticated models that can bridge the current gap between in vitro and in vivo worlds. Organs-on-chips, microphysiological systems, and AI-driven predictive modeling are emerging technologies that aim to provide the physiological relevance of in vivo studies at a lower cost and with higher throughput than traditional animal models [116] [114]. By integrating these advanced in vitro tools with targeted in vivo validation, researchers can build a more efficient, ethical, and predictive development pipeline, ultimately accelerating the delivery of new therapies to patients.
The journey of a biomaterial from the laboratory bench to regulatory approval hinges on a developer's ability to present a compelling, cohesive data package. This package must strategically integrate data from both in vitro (outside a living organism) and in vivo (within a living organism) studies to convincingly demonstrate safety and efficacy [117] [11]. For biomaterials, this is particularly critical, as a surprising poor correlation often exists between in vitro performance and in vivo outcomes [34]. A multicentre analysis revealed no significant overall correlation, with in vitro scores covarying with in vivo results only 58% of the time [34]. This article provides a guide for researchers on objectively comparing these testing results and building a data package that satisfies regulatory demands for groundbreaking medical devices.
Core Differences: Objective Comparison of In Vitro and In Vivo Models
Choosing between in vitro and in vivo models involves balancing control and complexity. The table below outlines their objective differences.
Table 1: Key Differences Between In Vitro and In Vivo Testing Models
| Aspect | In Vitro Models | In Vivo Models |
|---|---|---|
| Definition & Scope | Studies on isolated cells, tissues, or biomolecules in a controlled lab environment [117] [11] | Studies conducted within a whole, living organism [117] [11] |
| Physiological Relevance | Low; lacks systemic interactions between organs and biological systems [11] [110] | High; provides a holistic view of biological responses in a real-life context [117] [11] |
| Level of Control | High; enables the manipulation of specific variables in a simplified setup [117] [45] | Low; involves numerous unpredictable variables inherent to a living system [45] |
| Cost & Resources | Lower cost; requires fewer resources and materials [11] | High cost; involves animal care, monitoring, and extensive equipment [11] |
| Time to Results | Faster; allows for higher throughput and rapid screening [117] [11] | Longer; studies are extensive and time-consuming [11] |
| Ethical Considerations | Lower; generally does not involve live animals [11] | Significant ethical concerns; requires stringent oversight of animal use [117] [11] |
Quantitative Data Comparison: Analyzing Disparate Results
Understanding the quantitative performance gaps between testing models is essential for interpreting data. The following table summarizes potential comparative outcomes for a hypothetical bone regeneration biomaterial.
Table 2: Exemplary Quantitative Data Comparison for a Bone Regeneration Biomaterial
| Testing Parameter | In Vitro Results | In Vivo Results | Notes on Discrepancy |
|---|---|---|---|
| Cell Proliferation | 150% increase vs. control after 7 days [34] | 90% increase vs. control after 28 days [34] | The in vitro environment may oversupply growth factors, an effect moderated by systemic regulation in vivo. |
| Osteogenic Differentiation | 3.5-fold increase in osteocalcin expression [34] | 1.8-fold increase in bone volume fraction (BV/TV) [34] | Gene expression markers in vitro do not fully capture the complex, multi-cell type process of bone formation in vivo. |
| Angiogenic Potential | 2-fold increase in endothelial tube formation [118] | Moderate neovascularization, integration with host vasculature [118] | Co-culture systems in vitro cannot replicate the precise spatial and chemical gradients of angiogenic factors in a living organism. |
| Inflammatory Response | Low cytokine release (IL-6, TNF-α) in macrophage cultures [118] | Moderate, resolved acute inflammation; possible foreign body response [118] | In vitro assays often miss the sequential, multi-cellular cascade of the innate and adaptive immune response. |
| Degradation Rate | 5% mass loss in 4 weeks in buffer solution [34] | 15% mass loss in 4 weeks, mediated by enzymatic and cellular activity [34] | Static, enzymatic in vitro environments fail to mimic the dynamic and cellular-driven degradation in the body. |
Experimental Protocols: Core Methodologies for Biomaterial Testing
In Vitro Protocol: Cell Proliferation and Osteogenic Differentiation Assay
This protocol assesses a biomaterial's basic biocompatibility and capacity to promote bone-forming cell growth and differentiation [34].
- Material Extraction: Sterilize the biomaterial sample. Prepare an extract by incubing the material in cell culture medium (e.g., DMEM with 1% Penicillin-Streptomycin) for 24-72 hours at 37°C. Alternatively, seed cells directly onto 3D scaffolds [34].
- Cell Seeding: Use a relevant cell line, such as human Mesenchymal Stem Cells (hMSCs) or osteosarcoma-derived cells (e.g., MG-63). Seed cells in 24-well plates at a density of 1 x 10^4 cells per well, with or without the biomaterial extract or scaffold.
- Proliferation Assay (MTT):
- At defined time points (e.g., 1, 3, 7 days), add MTT reagent to each well and incubate for 3-4 hours.
- The metabolically active cells convert MTT to purple formazan crystals. Solubilize these crystals with a solvent like DMSO.
- Measure the absorbance at 570 nm using a plate reader. The absorbance is directly proportional to the number of viable cells [34].
- Osteogenic Differentiation:
- Culture cells in osteogenic induction medium (containing β-glycerophosphate, ascorbic acid, and dexamethasone) for 14-21 days.
- Quantitative Analysis: Quantify osteogenic markers:
- Alizarin Red S Staining: Fix cells and stain with Alizarin Red to detect calcium deposits. Quantify by eluting the stain and measuring absorbance.
- ALP Activity: Measure Alkaline Phosphatase (ALP) activity using a colorimetric assay with p-nitrophenyl phosphate as a substrate.
- Gene Expression: Perform RT-qPCR to measure the expression of osteogenic genes like RUNX2, OSTERIX, and OSTEOCALCIN [34].
In Vivo Protocol: Ectopic Bone Formation Model
This model evaluates a biomaterial's osteoinductive potential—its ability to stimulate de novo bone formation in a non-bony site [34].
- Animal Model & Implantation: Utilize an immunocompromised rodent model (e.g., NIH-III nude mouse). Anesthetize the animal according to approved ethical protocols.
- Make a small dorsal incision. Create subcutaneous pockets by blunt dissection.
- Implant the test biomaterial (e.g., a porous scaffold, often 5-8mm in diameter) into the pockets. Include both positive and negative control materials. Multiple implants can be placed per animal.
- Post-Operative Monitoring: Monitor animals for signs of pain, distress, or infection. Allow the implant to reside for a predetermined period, typically 4-8 weeks, to observe bone formation.
- Explant and Analysis: Euthanize the animal at the endpoint and surgically retrieve the implant with surrounding tissue.
- Histological Processing: Fix explants in formalin, dehydrate in ethanol, and embed in paraffin or resin. Section into thin slices (5-10 µm) and mount on slides.
- Staining and Imaging:
- Hematoxylin and Eosin (H&E): Provides a general overview of tissue architecture and cellular infiltration.
- Masson's Trichrome: Specifically stains collagen (blue/green) and can help identify newly formed osteoid matrix.
- Histomorphometry: Use image analysis software to quantify key parameters from the stained sections, such as the percentage of new bone area within the scaffold pores and the number of osteoclasts/osteoblasts at the material interface [34].
Visualizing the Testing and Submission Workflow
The path from initial testing to regulatory submission is a multi-stage, iterative process. The workflow diagram below outlines the key phases and decision points.
Figure 1: The iterative workflow for biomaterial testing and regulatory submission, highlighting the critical transition from in vitro to in vivo studies.
The Scientist's Toolkit: Essential Reagents for Biomaterial Testing
A successful testing strategy relies on specific reagents and tools. The following table details essential items for the featured experiments.
Table 3: Key Research Reagent Solutions for Biomaterial Testing
| Reagent / Material | Function in Experiment |
|---|---|
| Human Mesenchymal Stem Cells (hMSCs) | Primary cell model for assessing osteogenic differentiation potential and biocompatibility in vitro [34]. |
| Osteogenic Induction Medium | A specialized medium containing supplements (e.g., dexamethasone, β-glycerophosphate) to drive stem cells toward bone-forming osteoblasts [34]. |
| MTT Reagent Kit | A colorimetric assay for measuring cell proliferation and viability based on the reduction of yellow tetrazolium salt to purple formazan by metabolically active cells [34]. |
| PCR Primers for Osteogenic Markers | Specific primers for genes like RUNX2, OSTERIX, and OSTEOCALCIN are used in RT-qPCR to quantitatively measure differentiation at the genetic level [34]. |
| Immunodeficient Mouse Model | An in vivo model (e.g., NIH-III nude mouse) that prevents immune rejection of human-derived cells or materials, enabling the study of ectopic bone formation [34]. |
| Histology Staining Kits (H&E, Masson's Trichrome) | Essential for visualizing and analyzing tissue morphology, cellular response, and new matrix deposition on explanted biomaterials [34]. |
Navigating Regulatory Submission: From Data to Approval
A cohesive data package for a novel biomaterial must tell a compelling story to regulatory bodies like the FDA. The foundation is built on analytical studies that provide a detailed physical and chemical characterization of the material, demonstrating structural and functional similarity to a predicate device or its own design specifications [119]. The in vitro and in vivo data previously discussed form the core of the biological evaluation, addressing safety and performance.
Regulatory strategy should be integrated early in development. Engaging in early-stage consultations with regulatory agencies can help define the most appropriate preclinical testing approach, mitigate risks, and accelerate the path to market [120]. For products targeting serious conditions and unmet medical needs, expedited programs like the FDA's Breakthrough Therapy Designation (BTD) can be pursued. BTD requires preliminary clinical evidence indicating substantial improvement over existing therapies and offers benefits like more intensive FDA guidance [121].
The final submission should clearly explain how the in vitro data informed the in vivo study design and, crucially, how the in vivo results confirm the predictive value of the in vitro assays despite their inherent limitations. Acknowledging and rationalizing any discrepancies, rather than hiding them, strengthens the application's credibility [34] [110]. The ultimate goal is to present a weight-of-evidence package where all data streams converge to support the safety and efficacy of the biomaterial for its intended use.
Conclusion
The journey from laboratory concept to clinically successful biomaterial hinges on a sophisticated understanding of both in vitro and in vivo testing landscapes. Neither method alone can fully capture the complexity of the human body's response; rather, their true power is realized through strategic integration. The future of biomaterial testing lies in the continued development of more predictive in vitro systems that better mimic human physiology, such as advanced 3D models and organ-on-a-chip technologies, which align with the 3R principles to reduce animal testing. By adopting a holistic, iterative testing strategy that leverages the control of in vitro methods and the physiological relevance of in vivo studies, researchers can more efficiently de-risk development, accelerate innovation, and ultimately deliver safer, more effective biomaterials to patients.
References
- 1. In vitro [https://en.wikipedia.org/wiki/In_vitro]
- 2. In vivo vs. in vitro: What is the difference? [https://www.medicalnewstoday.com/articles/in-vivo-vs-in-vitro]
- 3. In Vivo Study - an overview [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/in-vivo-study]
- 4. Understanding In Vivo vs In Vitro Methods [https://www.technologynetworks.com/drug-discovery/articles/in-vivo-vs-in-vitro-definition-pros-and-cons-350415]
- 5. In Vitro to In Vivo Translation in Lead Optimization [https://www.sygnaturediscovery.com/news-and-events/blog/in-vitro-to-in-vivo-translation-in-lead-optimization/]
- 6. Complex in vitro model: A transformative model in drug ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10828975/]
- 7. Predicting In Vivo Responses to Biomaterials via Combined In ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4313398/]
- 8. Comparing in vitro, in situ, and in vivo experimental data ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC22353/]
- 9. Advanced testing and biocompatibility strategies for ... [https://biotechsustainablematerials.biomedcentral.com/articles/10.1186/s44316-024-00018-7]
- 10. In vitro or In vivo. What is the difference? [https://www.ainia.com/en/ainia-news/in-vitro-or-in-vivo-what-is-the-difference/]
- 11. In Vivo vs. In Vitro Models: Key Differences and ... [https://liveonbiolabs.com/in-vivo-vs-in-vitro-models/]
- 12. In Vitro vs In Vivo: A History of Modern Cell Culture [https://emulatebio.com/in-vitro-vs-in-vivo-cell-culture/]
- 13. A Review of Biomaterials and Associated Performance ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9607025/]
- 14. Global Biomaterial Testing Equipment Market Size, Share, ... [https://www.databridgemarketresearch.com/reports/global-biomaterial-testing-equipment-market?srsltid=AfmBOop5S74nJKaAmm8puvMErXOYeyOoH12M8V7q1x31FcYfaSARYVEq]
- 15. Novel 'living' biomaterial aims to advance regenerative ... [https://www.sciencedaily.com/releases/2025/02/250206155347.htm]
- 16. Navigating Biocompatibility: An Introduction to ISO 10993 ... [https://medinstitute.com/blog/navigating-biocompatibility-an-introduction-to-iso-10993-series-of-standards/]
- 17. Use of International Standard ISO 10993-1, "Biological ... [https://www.fda.gov/regulatory-information/search-fda-guidance-documents/use-international-standard-iso-10993-1-biological-evaluation-medical-devices-part-1-evaluation-and]
- 18. ISO 10993-1:2018 - Biological evaluation of medical devices [https://www.iso.org/standard/68936.html]
- 19. ISO 10993 [https://en.wikipedia.org/wiki/ISO_10993]
- 20. ISO 10993-1:2025 Updates —Top 10 Biological Evaluation ... [https://namsa.com/resources/blog/iso-10993-12025top-10-biological-evaluation-essentials-in-the-update/]
- 21. The “Big Three” in biocompatibility testing of medical devices [https://pmc.ncbi.nlm.nih.gov/articles/PMC10800850/]
- 22. Biocompatibility & Sterilization in 3D Printed Medical Devices | ISO ... [https://www.stratasys.com/en/stratasysdirect/resources/articles/biocompatibility-sterilization-3d-printed-medical-devices/]
- 23. From in vitro Experiments to in vivo and Clinical Studies [https://pubmed.ncbi.nlm.nih.gov/26778084/]
- 24. How does in vitro testing compare with in vivo testing? [https://www.certisoncology.com/faqs/how-does-in-vitro-testing-compare-with-in-vivo-testing/]
- 25. In Vivo vs. In Vitro: Advantages & Disadvantages [https://www.medicinenet.com/why_is_in_vivo_better_than_in_vitro/article.htm]
- 26. Improved analysis of in vivo drug combination experiments ... [https://www.nature.com/articles/s41467-025-65218-9]
- 27. Principles and Strengths of In Vitro Testing [https://www.eag.com/blog/principles-and-strengths-of-in-vitro-testing/]
- 28. In vitro test systems and their limitations - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4803002/]
- 29. The merits of in vitro versus in vivo modeling ... [https://www.sciencedirect.com/science/article/abs/pii/S1382668905001158]
- 30. A review of cytotoxicity testing methods and in vitro study of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12534221/]
- 31. Summary of Advantages and Disadvantages of In Vitro ... - NCBI [https://www.ncbi.nlm.nih.gov/books/NBK100200/]
- 32. In Vitro Testing in Cosmetics: Pros and Cons [https://qima-lifesciences.com/cosmetic_publication/in-vitro-testing-pros-cons/]
- 33. Investigating Immunomodulatory Biomaterials for ... [https://www.mdpi.com/2306-5354/10/12/1411]
- 34. A surprisingly poor correlation between in vitro and in vivo ... [https://pubmed.ncbi.nlm.nih.gov/27215739/]
- 35. Comparative Evaluation of Biocompatibility Data from the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12588249/]
- 36. In vitro and in vivo assessment of a new acellular human ... [https://www.nature.com/articles/s41598-025-88814-7]
- 37. Exploration of biomaterial-tissue integration in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11968225/]
- 38. In Vitro vs. In Vivo: What's the Difference? [https://newlifefertility.com/blog/in-vitro-and-in-vivo/]
- 39. In Vitro Hemocompatibility and Genotoxicity Evaluation of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9959965/]
- 40. Cytotoxicity, Sensitization & Irritation Testing Explained [https://nabi.bio/cytotoxicity-sensitization-and-irritation-testing-what-do-they-involve/]
- 41. Proof of concept testing of a positive reference material for ... [https://pubmed.ncbi.nlm.nih.gov/38334720/]
- 42. Determination of Chemical Irritation Potential Using a ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8659397/]
- 43. A database of human predictive patch test data for skin ... [https://link.springer.com/article/10.1007/s00204-023-03530-3]
- 44. In vitro and in vivo biodegradation and biocompatibility ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2025.1685918/full]
- 45. . In Trials (and Why Combining Both...) | HowStuffWorks Vivo vs In Vitro [https://science.howstuffworks.com/life/cellular-microscopic/in-vivo-vs-in-vitro.htm]
- 46. selection of appropriate animal models - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9255784/]
- 47. Neurogenesis in primates versus rodents and the value of non ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10659238/]
- 48. Regulatory considerations for animal studies of biomaterial ... [https://www.sciencedirect.com/science/article/pii/S2452199X21004497]
- 49. A Review on Biomaterials for Orthopaedic Surgery and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9145924/]
- 50. Recent advances in regenerative biomaterials - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9745784/]
- 51. Quality and Utility of European Cardiovascular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10702370/]
- 52. Tissue Engineered Materials in Cardiovascular Surgery [https://www.frontiersin.org/journals/cardiovascular-medicine/articles/10.3389/fcvm.2020.00055/full]
- 53. Comparison of Printable Biomaterials for Use in Neural ... [https://www.mdpi.com/2073-4360/16/10/1426]
- 54. The Role of Artificial Intelligence in Biomaterials Science [https://www.mdpi.com/2073-4360/17/19/2668]
- 55. Future challenges in the in vitro and in vivo evaluation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4817327/]
- 56. Discordance between histologic and visual assessment of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6393178/]
- 57. Automated cell annotation and classification on ... [https://www.nature.com/articles/s41467-025-61349-1]
- 58. Understanding In Vitro & In Vivo Testing [https://www.novology.com/blogs/dry-skin/in-vivo-in-vitro-difference?srsltid=AfmBOorh4TMqGNLJy-4fAaM8kxNqiI_TMyeaPg6zRbjCug3AxTHtZZqq]
- 59. In Vitro vs In Vivo: Complete Comparison + Selection Guide [https://www.assaygenie.com/in-vitro-vs-in-vivo-complete-comparison-selection-guide-research-methods/?srsltid=AfmBOoplm-A520A74epYLtyNkIjrquLeNoQCOyrzfO3zQsY0hf9rVMpH]
- 60. Why 90% of clinical drug development fails and how to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9293739/]
- 61. The Reality of Drug Discovery and Development [https://humanspecificresearch.org/the-harsh-reality-of-drug-discovery-and-development/]
- 62. In Vivo Research Strategies that Reduce Risk of Failure ... [https://www.criver.com/eureka/vivo-research-strategies-reduce-risk-failure-clinic]
- 63. In Vitro vs. In Vivo vs. In Silico: Preclinical Assay Differences [https://www.zeclinics.com/blog/differences-between-in-vitro-in-vivo-and-in-silico-assays-in-preclinical-research]
- 64. Understanding the correlation between in vitro and in vivo ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5831149/]
- 65. What Is IVIVE? 7 FAQs About In Vitro-In Vivo Extrapolation [https://labtesting.wuxiapptec.com/2025/09/11/in-vitro-in-vivo-extrapolation-what-developers-should-know/]
- 66. AIVIVE: a novel AI framework for enhanced in vitro to in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12469192/]
- 67. Special Issue : In Vitro–In Vivo Correlation and Mechanistic ... [https://www.mdpi.com/journal/pharmaceutics/special_issues/Z913E25BJ6]
- 68. Level B and C in vivo/in vitro correlations: statistical ... [https://www.sciencedirect.com/science/article/abs/pii/S0378517397002627]
- 69. Human disease models in drug development [https://www.nature.com/articles/s44222-023-00063-3]
- 70. Three-Dimensional Culture System: A New Frontier in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12292892/]
- 71. 3D Cell Culture Methods and Applications [https://elveflow.com/microfluidic-reviews/3d-cell-culture-methods-and-applications-a-short-review/]
- 72. The Development of 3D Primary Co-Culture Models ... [https://pubmed.ncbi.nlm.nih.gov/40507840/]
- 73. Human Primary Cell Culture Market Size & Forecast, 2025 ... [https://www.coherentmarketinsights.com/industry-reports/human-primary-cell-culture-market]
- 74. 3d Cell Culture Market: Comprehensive Analysis and ... [https://www.pharmiweb.com/press-release/2025-02-12/3d-cell-culture-market-comprehensive-analysis-and-forecast-2025-2035]
- 75. Using in vitro human disease cell models in drug discovery [https://www.drugtargetreview.com/article/105164/right-model-right-complexity-using-in-vitro-human-disease-cell-models-in-drug-discovery/]
- 76. Blood Plasma, Fibrinogen or Fibrin Biomaterial for the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11942893/]
- 77. Blood-derived biomaterials for tissue graft biofabrication by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10339460/]
- 78. Dynamic Culture Conditions to Generate Silk-Based Tissue ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2732107/]
- 79. Dynamic culture enhances stem cell infiltration and ... [https://www.sciencedirect.com/science/article/abs/pii/S1742706110003818]
- 80. In vitro evaluation of blood plasma coagulation responses ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10408244/]
- 81. A mechanistic model of in vitro plasma activation to evaluate ... [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1012552]
- 82. Biocompatibility assessment of biomaterials used in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8461597/]
- 83. Macrophages in organ fibrosis: from pathogenesis to ... [https://www.nature.com/articles/s41420-024-02247-1]
- 84. Macrophage heterogeneity in liver fibrosis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12443557/]
- 85. The impact of biomaterial characteristics on macrophage ... [https://www.oaepublish.com/articles/2347-9264.2024.166]
- 86. Towards predicting implant-induced fibrosis: A standardized ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12312070/]
- 87. Understanding fibrosis pathogenesis via modeling ... [https://www.nature.com/articles/s41467-022-34241-5]
- 88. A novel ex vivo approach for investigating profibrotic ... [https://www.sciencedirect.com/science/article/pii/S0006291X24015742]
- 89. Pexidartinib and Nintedanib Combination Therapy Targets ... [https://www.mdpi.com/1422-0067/26/15/7570]
- 90. The Role of Artificial Intelligence in Biomaterials Science [https://pmc.ncbi.nlm.nih.gov/articles/PMC12526772/]
- 91. Computational modelling of biological systems now and then [https://pmc.ncbi.nlm.nih.gov/articles/PMC12105733/]
- 92. While AI Could Be the Game Changer in Predicting Health ... [https://www.medschool.umaryland.edu/news/2025/while-ai-could-be-the-game-changer-in-predicting-health-outcomes-it-should-not-be-the-only-method.html]
- 93. Application of Machine Learning in Predicting Osteogenic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12561851/]
- 94. Practical Perspectives on Enhancing Predictive Modeling ... [https://link.springer.com/article/10.1007/s40495-025-00426-x]
- 95. Computational methods and artificial intelligence-based ... [https://www.frontiersin.org/journals/materials/articles/10.3389/fmats.2025.1645227/full]
- 96. 5E: Computational and Machine Learning Approaches for ... [https://2025.biomaterials.org/program/sessions/5e-computational-and-machine-learning-approaches-biomaterial-design-evaluation]
- 97. How artificial intelligence shapes the future of biomaterials? [https://www.sciencedirect.com/science/article/pii/S2949822824002788]
- 98. In vitro and in vivo comparison of five biomaterials used for ... [https://pubmed.ncbi.nlm.nih.gov/18167101/]
- 99. Comparison of mechanical testing methods for biomaterials [https://www.sciencedirect.com/science/article/abs/pii/S1751616115002635]
- 100. In Vitro Toxicity Testing Market Size & Forecast, 2025-2032 [https://www.coherentmarketinsights.com/market-insight/in-vitro-toxicity-testing-market-3855]
- 101. In Vivo Toxicology Market Size, Share & Trends [2025] [https://www.marketsandmarkets.com/Market-Reports/in-vivo-toxicology-testing-market-105308811.html]
- 102. Making in vitro release and formulation data AI-ready [https://pmc.ncbi.nlm.nih.gov/articles/PMC12481109/]
- 103. A machine learning workflow to accelerate the design of in ... [https://pubs.rsc.org/en/content/articlehtml/2025/dd/d5dd00112a]
- 104. Can acute oral in vivo toxicity testing for EU REACH be ... [https://www.sciencedirect.com/science/article/abs/pii/S0273230025000911]
- 105. Advancing biomaterial research with artificial intelligence [https://www.sciencedirect.com/science/article/abs/pii/S2772950825003620]
- 106. Advances in Biodegradable Polymers and Biomaterials for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10488517/]
- 107. Validation of the Chick Chorioallantoic Membrane (CAM) ... [https://iv.iiarjournals.org/content/39/3/1740]
- 108. In vitro calibration and in vivo validation of ... [https://www.sciencedirect.com/science/article/pii/S1742706124001612]
- 109. Auxetic Meta-Biomaterials: Computer Simulation and ... [https://www.mdpi.com/2075-4701/15/3/241]
- 110. In vitro vs. In vivo: Is One Better? | UHN Research [https://www.uhnresearch.ca/news/vitro-vs-vivo-one-better]
- 111. The Value of Systematic Reviews in Estimating the Cost ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5124733/]
- 112. In Vitro vs In Vivo: Complete Comparison + Selection Guide [https://www.assaygenie.com/in-vitro-vs-in-vivo-complete-comparison-selection-guide-research-methods/?srsltid=AfmBOor7HbN2TRFRpQpPdgUC-9sA0-XQNuzuBRT2XX1eo9kORk_NyMKI]
- 113. Improving In Vitro–In Vivo Correlation (IVIVC) for Lipid ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12567226/]
- 114. CMC Perspectives of In Vitro / In Vivo Correlation (IVIVC) ... [https://premier-research.com/perspectives/cmc-perspectives-of-in-vitro-in-vivo-correlation-ivivc-in-drug-development/]
- 115. Ethical considerations on the moral status of the embryo and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11532601/]
- 116. Global Biomaterial Testing Equipment Market Analysis ... [https://www.linkedin.com/pulse/global-biomaterial-testing-equipment-market-analysis-2025-2032-r2src]
- 117. In Vitro vs In Vivo: Complete Comparison + Selection Guide [https://www.assaygenie.com/in-vitro-vs-in-vivo-complete-comparison-selection-guide-research-methods/?srsltid=AfmBOoqn6QHPOeoXFEQJTwrPVvek-mCjAPCrSEZwLR_Sp94zueQy3ZSj]
- 118. In vitro and in vivo interactions of cells with biomaterials [https://www.sciencedirect.com/science/article/pii/0142961288900634]
- 119. Review and Approval [https://www.fda.gov/drugs/biosimilars/review-and-approval]
- 120. From Lab to Launch: Ensuring Regulatory Compliance for ... [https://ifyber.com/from-lab-to-launch-ensuring-regulatory-compliance-for-groundbreaking-medical-devices/]
- 121. Introduction to Expedited Regulatory Pathways - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7214660/]