Evidence Levels in Biomaterial Research: A Framework from Bench to Bedside
This article provides a comprehensive framework for understanding the hierarchy of evidence in biomaterials research, tailored for researchers, scientists, and drug development professionals.
Evidence Levels in Biomaterial Research: A Framework from Bench to Bedside
Abstract
This article provides a comprehensive framework for understanding the hierarchy of evidence in biomaterials research, tailored for researchers, scientists, and drug development professionals. It explores the foundational concepts of evidence-based biomaterials, detailing the translation roadmap from basic research to commercial medical products. The content delves into methodological applications, including systematic reviews and meta-analyses, for generating robust scientific evidence. It further addresses common challenges in study design and data interpretation, offering optimization strategies and exploring the role of advanced tools like AI. Finally, it covers validation and comparative techniques essential for regulatory approval and clinical translation, synthesizing key takeaways to guide future research and development in the field.
What Are Evidence Levels? Foundational Principles for Biomaterial Scientists
Defining the EBBR Framework in Preclinical Research
Evidence-Based Biomaterials Research (EBBR) represents a systematic approach to evaluating biomaterial efficacy and safety through rigorously designed preclinical studies. This methodology emphasizes standardized experimental parameters, appropriate model selection, and robust data analysis to generate reliable, translatable evidence for clinical applications. In joint repair research, for instance, EBBR principles have been applied to evaluate a plethora of promising devices, with the recognition that study design significantly influences therapeutic outcomes and clinical translation potential [1]. The framework provides a structured pathway for navigating the complex journey from laboratory discovery to clinical implementation, addressing the critical need for consensus in experimental design that has historically made conclusions on efficacy difficult to draw [1].
The foundation of EBBR rests upon eliminating potential sources of bias through meticulous experimental planning. Key bias domains addressed in rigorous EBBR include selection bias (through proper randomization), detection bias (via blinded outcome assessment), attrition bias (by accounting for all results), and reporting bias (through comprehensive outcome reporting) [1]. This systematic approach to bias management enhances the reliability of preclinical findings, which is essential for successful progression through the stringent review processes required for regulatory approval [1]. By implementing these principles, EBBR aims to standardize experimental approaches across the field, ultimately advancing the effective translation of novel biomaterial-based therapeutics to the clinic.
Core Methodological Principles of EBBR
Systematic Literature Review and Risk of Bias Assessment
The EBBR approach employs systematic literature review methodologies to identify key experimental parameters that significantly affect biomaterial therapeutic outcomes. This process begins with defined search criteria across scientific databases such as PubMed, using specific terms related to the biomaterial application [1]. For joint repair studies, inclusion criteria typically focus on English-language publications utilizing large animal models (pig, goat, sheep, horse) investigating biomaterial-based treatments for knee cartilage defects, while excluding studies incorporating growth factors, genetic material, or other biological fluids [1].
Risk of Bias (RoB) assessment forms the cornerstone of EBBR quality evaluation. Modified tools like the Office of Health Assessment and Translation (OHAT) and Systematic Review Centre for Laboratory Animal Experimentation (SYRCLE) are utilized to score studies across multiple bias domains [1]. Studies are evaluated based on sample allocation sequences, blinded outcome assessment, completeness of outcome reporting, use of appropriate validated outcomes, ethical approval, and conflict of interest declarations [1]. This rigorous scoring system allows researchers to identify high-quality studies with low RoB, which form the evidence base for reliable conclusions about biomaterial performance.
Key Experimental Parameters in Biomaterial Evaluation
EBBR emphasizes the critical importance of standardizing key experimental parameters to enable valid comparisons between studies. Through systematic analysis of low RoB publications, EBBR has identified several factors that significantly influence outcomes in joint repair research, including defect localization, animal age and maturity, selection of appropriate controls, and the use of cell-free versus cell-laden biomaterials [1].
The selection of animal models represents another crucial consideration in EBBR. Different species offer distinct advantages and limitations; for example, miniature pigs are often preferred due to joint size and cartilage thickness (0.5-1.5 mm in the medial femoral condyle) similar to humans (1.69-2.55 mm), while sheep models show more variability in cartilage thickness (0.45-1.68 mm) that can lead to inconsistent outcomes [1]. Understanding these model-specific characteristics is essential for proper study design and interpretation of results within the EBBR framework.
Comparative Analysis of Biomaterial Performance
EBBR Application in Osteochondral Repair
The systematic application of EBBR principles to osteochondral repair studies has yielded significant insights into biomaterial performance. Analysis of low RoB studies revealed that mechanically strong biomaterials perform better at the femoral condyles, while highlighting the importance of including native tissue controls to properly evaluate the quality of newly formed tissue [1]. Furthermore, in cell-laden biomaterials, the pre-culture conditions played a more important role in defect repair than the cell type itself [1].
Table 1: Key Experimental Parameters Identified Through EBBR in Osteochondral Repair Studies
| Parameter Category | Specific Factors | Impact on Outcomes |
|---|---|---|
| Defect Characteristics | Location (femoral condyle vs. other sites) | Mechanically strong biomaterials perform better at femoral condyles [1] |
| Size and volume | Must be standardized relative to species-specific cartilage thickness [1] | |
| Animal Model | Species selection (pig, sheep, goat, horse) | Influences comparability to human pathology and healing processes [1] |
| Age and maturity | Affects healing response and translation to human clinical scenarios [1] | |
| Experimental Design | Control group selection | Native tissue controls essential for proper evaluation of repair tissue quality [1] |
| Cell-free vs. cell-laden approaches | Cell pre-culture conditions more critical than cell type [1] |
These findings demonstrate how EBBR methodologies can extract meaningful patterns from diverse studies, providing evidence-based guidance for future research directions and clinical translation strategies. The systematic comparison approach allows researchers to identify not only which biomaterials show promise, but also under what specific conditions they demonstrate efficacy.
Natural vs. Synthetic Biomaterials in Cardiac Tissue Engineering
EBBR principles have been applied to compare natural and synthetic biomaterials for cardiac tissue regeneration, revealing distinct advantages and limitations for each category. Natural biomaterials like chitin, chitosan, cellulose, and hyaluronic acid demonstrate excellent biodegradability and biocompatibility with human cardiac tissue [2]. In contrast, synthetic biomaterials such as poly-vinyl alcohol, polyglycolic acid, and polyurethane offer high customizability but may lack the innate biological recognition of natural alternatives [2].
The EBBR approach indicates that natural biomaterials currently fulfill more desired characteristics for cardiac tissue regeneration compared to synthetic options [2]. However, the framework also highlights the emerging potential of hybrid biomaterials that combine advantages from both categories, suggesting this may represent the optimal solution for future therapeutic development [2]. This evidence-based comparison provides valuable guidance for researchers selecting biomaterial strategies for specific cardiac repair applications.
Table 2: Comparative Analysis of Biomaterial Categories Through EBBR Framework
| Evaluation Criterion | Natural Biomaterials | Synthetic Biomaterials | Hybrid Approach |
|---|---|---|---|
| Biocompatibility | Excellent compatibility with human cardiac tissue [2] | Variable; depends on material composition and degradation products | Can be engineered to optimize compatibility |
| Biodegradability | Controlled, predictable degradation profiles [2] | Tunable but may produce less biocompatible breakdown products | Balance of natural degradation with synthetic control |
| Customizability | Limited by natural material properties | High degree of chemical and physical customization [2] | Maximum design flexibility combining both approaches |
| Mechanical Properties | May lack required strength for some applications | Highly tunable mechanical characteristics [2] | Can achieve ideal mechanical match for target tissue |
| Clinical Translation | Generally favorable safety profiles | May face regulatory hurdles due to novel chemistry | Combines established natural materials with enhanced functionality |
Experimental Protocols in EBBR
Large Animal Model Selection and Surgical Protocols
EBBR emphasizes standardized surgical protocols for creating osteochondral defects in large animal models. The recommended approach involves creating critical-sized defects in weight-bearing areas of the femoral condyles, with dimensions proportional to the species-specific joint anatomy [1]. For miniature pigs, defect diameter typically ranges from 4-6 mm, while in sheep models, 4-5 mm defects are common, reflecting the smaller joint dimensions [1]. These defect sizes must be calibrated against the specific cartilage thickness of the chosen model, which varies significantly between species - from 0.45-1.68 mm in sheep to 0.5-1.5 mm in miniature pigs [1].
Postoperative care and evaluation timelines represent another critical component of EBBR protocols. Studies employing EBBR principles typically include standardized rehabilitation protocols, pain management strategies, and predetermined evaluation timepoints (e.g., 3, 6, and 12 months) to assess both short-term and long-term outcomes [1]. The inclusion of multiple assessment timepoints allows for evaluation of the temporal progression of tissue integration, remodeling, and maturation, providing a more comprehensive understanding of biomaterial performance throughout the healing process.
Outcome Assessment Methodologies
EBBR incorporates comprehensive, multimodal outcome assessments to evaluate biomaterial efficacy. Macroscopic evaluation includes scoring systems for tissue appearance, integration with surrounding tissue, and surface characteristics [1]. Histological analyses provide critical information about tissue architecture, cellular distribution, and extracellular matrix composition using standardized staining protocols (e.g., hematoxylin and eosin, safranin-O, collagen type II immunohistochemistry) [1].
Biomechanical testing forms another essential component of EBBR assessment protocols, evaluating the functional integration of the repair tissue through indentation testing, tensile testing, and other mechanical property assessments [1]. These quantitative measurements provide crucial data about the functional performance of the regenerated tissue compared to native cartilage. Additionally, biocompatibility evaluations assess local and systemic inflammatory responses, degradation characteristics of the biomaterial, and overall tissue biocompatibility through histological analysis and evaluation of immune cell infiltration [1].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Essential Research Reagents and Materials for EBBR Studies
| Reagent/Material | Function/Application | Specific Examples |
|---|---|---|
| Natural Biomaterials | Provide biocompatible scaffolding with inherent biological recognition | Chitin, chitosan, cellulose, hyaluronic acid [2] |
| Synthetic Biomaterials | Offer highly customizable mechanical and chemical properties | Poly-vinyl alcohol, polyglycolic acid, polyurethane [2] |
| Cell Sources | Provide cellular component for cell-laden approaches | Chondrocytes, mesenchymal stem cells, induced pluripotent stem cells [1] |
| Histological Stains | Enable evaluation of tissue architecture and composition | Hematoxylin and eosin, safranin-O, collagen type II immunohistochemistry [1] |
| Biomechanical Testing Equipment | Assess functional integration and mechanical properties | Indentation testers, tensile testing systems, dynamic mechanical analyzers [1] |
| HEX azide, 6-isomer | HEX azide, 6-isomer, MF:C24H12Cl6N4O6, MW:665.1 g/mol | Chemical Reagent |
| 2-Fluoro-5-iodobenzylamine | 2-Fluoro-5-iodobenzylamine, 771572-96-4 | 2-Fluoro-5-iodobenzylamine (CAS 771572-96-4) is a key building block for pharmaceutical and chemical research. For Research Use Only. Not for human use. |
This toolkit represents the essential components for conducting rigorous EBBR studies in tissue engineering and regenerative medicine. The selection of specific reagents and materials should be guided by the research question, target tissue, and desired outcomes, with careful consideration of the evidence supporting each component's efficacy for the intended application.
The EBBR framework represents a paradigm shift in biomaterial evaluation, emphasizing evidence-based decision-making throughout the research process. By implementing systematic methodologies, standardized protocols, and comprehensive outcome assessments, EBBR enhances the reliability, reproducibility, and clinical translatability of biomaterial research findings. The comparative analyses enabled by this approach provide valuable insights into the performance characteristics of different biomaterial strategies under specific experimental conditions, guiding future research directions and clinical application decisions.
As the field continues to evolve, EBBR principles will play an increasingly important role in navigating the complex landscape of biomaterial development and validation. The continued refinement of EBBR methodologies, including the development of more sophisticated outcome measures and standardized reporting guidelines, will further enhance the quality and impact of biomaterial research. Ultimately, the widespread adoption of EBBR principles promises to accelerate the translation of promising biomaterial technologies from the laboratory to clinical practice, addressing unmet needs in tissue repair and regeneration.
The development of biomaterials from basic research to commercialized medical products is a complex, multidisciplinary endeavor fraught with translational obstacles. The biomaterials market, estimated at over 100 billion USD in 2019 with a compound annual growth rate of 15.9%, demonstrates significant therapeutic and commercial potential, yet the journey from discovery to clinical adoption often takes decades [3]. For instance, the translation of PLGA from initial biocompatibility studies to an approved product for prostate cancer therapy spanned approximately 20 years [3]. This lengthy timeline underscores the critical need for robust methodologies to validate and prioritize promising biomaterials research.
The emerging discipline of Evidence-Based Biomaterials Research (EBBR) addresses this need by applying systematic, evidence-based approaches—spearheaded by systematic reviews and meta-analyses—to translate vast research data into validated scientific evidence [4] [5]. This paradigm shift moves the field beyond unstructured empirical approaches toward development strategies entrenched in data validation, enabling researchers to navigate the "valley of death" where promising academic solutions frequently stall [6]. This guide compares traditional and evolving research strategies within a hierarchical evidence framework, providing researchers with a structured approach to evaluate biomaterials' clinical potential.
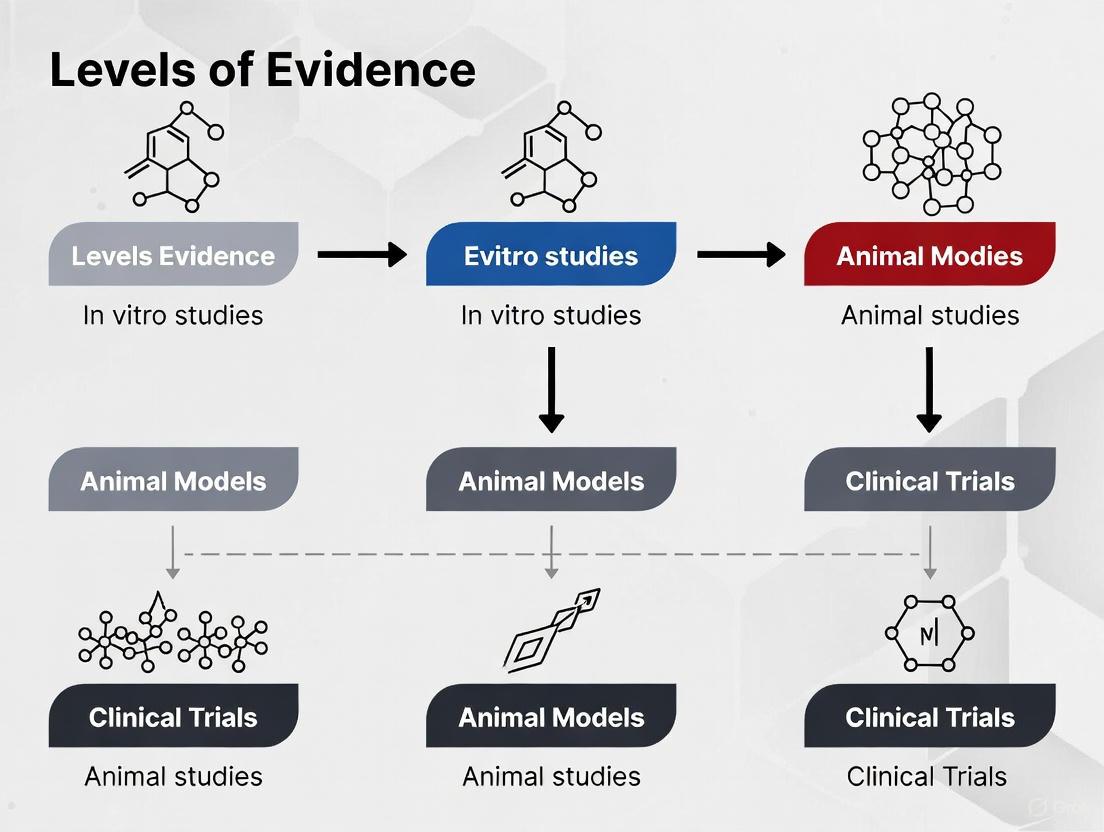
Levels of Evidence in Biomaterials Research
In evidence-based practice, research findings are categorized according to their inherent reliability and freedom from bias. This hierarchy, illustrated below, provides a critical framework for assessing the strength of biomaterials research claims.
Figure 1: The Evidence Hierarchy Pyramid for Biomaterials Research. Level I represents the strongest, most synthesized evidence, while Level V represents the least filtered information. Adapted from established evidence-based medicine principles [7] [8].
Understanding the Evidence Levels
- Level I (Strongest Evidence): Systematic reviews and meta-analyses comprehensively synthesize all available evidence on a specific research question. For example, an EBBR approach using systematic review can generate validated evidence about a biomaterial's performance across multiple studies, providing the highest confidence for clinical decision-making [4] [7].
- Level II: Randomized controlled trials (RCTs) provide high-quality experimental evidence where participants are randomly allocated to treatment or control groups. While more common in clinical settings, controlled preclinical studies in animal models also fall under this category when properly randomized [7] [8].
- Level III: Cohort studies follow groups of subjects (e.g., animals or human patients) exposed to a particular biomaterial over time, comparing their outcomes to unexposed groups. These observational studies provide valuable longitudinal data but with less control than RCTs [7] [9].
- Level IV: Case-control studies and case series report on outcomes for a small group of subjects or a single subject without control groups. These are often used for initial safety reports or preliminary efficacy signals [7] [8].
- Level V (Weakest Evidence): Expert opinion, background information, and mechanistic studies based on non-systematic reviews represent the lowest level of evidence, though they can provide valuable foundational knowledge for further research [7] [8].
Comparative Analysis of Biomaterial Research Approaches
Traditional Empirical Development vs. Evidence-Based and Data-Driven Approaches
The biomaterials field is undergoing a fundamental transformation from reliance on traditional empirical methods to more systematic, evidence-based approaches. The table below compares these evolving research paradigms.
Table 1: Comparison of Biomaterials Research and Development Approaches
| Research Aspect | Traditional Empirical Approach | Evidence-Based Approach (EBBR) | Data-Driven/Machine Learning Approach |
|---|---|---|---|
| Core Methodology | Trial-and-error experimentation based on researcher intuition and literature precedents [3] | Systematic review and meta-analysis of existing research data [4] [5] | Machine learning algorithms trained on high-throughput experimental data [3] |
| Data Utilization | Limited, unstructured data analysis; focus on individual studies | Comprehensive synthesis of all available published data | Analysis of multi-dimensional datasets including composition, structure, and properties [3] |
| Translation Timeline | Lengthy (e.g., ~20 years for PLGA translation) [3] | Potentially accelerated through validated evidence | Dramatically accelerated prediction of optimized materials (e.g., 10 hours vs. 5 days for composites) [3] |
| Key Strengths | Direct researcher control; established protocols | High-quality, validated evidence; reduced bias | Handles complex parameter spaces; discovers non-intuitive relationships [3] |
| Limitations | Susceptible to bias; limited scope; inefficient | Dependent on quality of primary studies | Requires large, high-quality datasets; "black box" limitations |
Case Comparison: Printable Biomaterials for Neural Tissue Engineering
Recent research on neural tissue engineering scaffolds provides exemplary data for comparing biomaterials across multiple evidence levels. The following table synthesizes experimental findings from a comprehensive study evaluating four thermoplastics and one hydrogel for neural applications [10].
Table 2: Experimental Comparison of 3D-Printed Biomaterials for Neural Tissue Engineering
| Biomaterial | Printability & Resolution | Mechanical Properties | In Vitro Cell Viability | In Vivo Biocompatibility (10 days) |
|---|---|---|---|---|
| PLA | Superior printability, high resolution and shape fidelity [10] | Excellent thermal stability, degradability [10] | Moderate | Connective tissue encapsulation with some inflammatory cells [10] |
| PCL | Superior printability, high resolution and shape fidelity [10] | Mechanical elasticity, long-term degradation, low melting point [10] | Moderate | Connective tissue encapsulation with some inflammatory cells [10] |
| Filaflex (FF) | Good printing results [10] | High flexibility (92A shore hardness), electroconductive [10] | Moderate | Connective tissue encapsulation with some inflammatory cells [10] |
| Flexdym (FD) | Good printing results [10] | Great viscoelastic properties, flexible, stretchable [10] | Moderate | Connective tissue encapsulation with some inflammatory cells [10] |
| GelMA Hydrogel | Lower resolution compared to thermoplastics [10] | Great viscoelastic properties, tunable mechanical properties [10] | Greater cell viability after 7 days [10] | Connective tissue encapsulation with some inflammatory cells [10] |
This comparative data demonstrates how different biomaterials offer distinct advantages depending on the target application requirements, with thermoplastics generally providing superior structural characteristics while hydrogels like GelMA offer enhanced biocompatibility.
Experimental Protocols for Biomaterial Evaluation
Standardized Workflow for Biomaterial Characterization
The translation of biomaterials requires rigorous, standardized characterization across multiple domains. The following diagram outlines a comprehensive experimental workflow for evaluating candidate biomaterials.
Figure 2: Progressive Workflow for Biomaterial Evaluation. The workflow progresses from material characterization (yellow) to in vitro testing (blue) and finally to in vivo assessment (red/green), representing increasing evidence levels [10].
Detailed Methodologies for Key Experiments
Printability and Mechanical Testing Protocol
Objective: Evaluate biomaterials for extrusion-based 3D printing and characterize mechanical properties relevant to neural tissue engineering [10].
Materials Preparation:
- Thermoplastics: Use ready-to-use filaments (PLA, PCL, Filaflex) or pellets (Flexdym) [10].
- Hydrogel: Prepare 10% GelMA/0.5 LAP (w/v) by dissolving 60 mg LAP (lithium phenyl-2,4,6-trimethylbenzoylphosphinate) in 12 mL of 1× sterile phosphate buffer solution, then adding lyophilized GelMA [10].
Printability Assessment:
- Utilize extrusion-based 3D printing systems (e.g., fused deposition modeling for thermoplastics, 3D plotting for hydrogels).
- Assess printing parameters: resolution, shape fidelity, structural integrity.
- Optimize nozzle temperature (for thermoplastics) or crosslinking conditions (for photopolymerizable hydrogels like GelMA) [10].
Mechanical Testing:
- Perform uniaxial tensile testing according to ASTM D638 standards.
- Measure elastic modulus, ultimate tensile strength, and elongation at break.
- For viscoelastic materials (e.g., FD, GelMA), conduct dynamic mechanical analysis to characterize storage and loss moduli [10].
In Vitro Cell-Biomaterial Interaction Protocol
Objective: Evaluate cellular response to biomaterials, including viability, adhesion, and proliferation [10].
Cell Culture:
- Use relevant cell lines (e.g., neural stem cells, Schwann cells) or primary cells.
- Culture cells according to established protocols with appropriate media supplements.
- Seed cells onto sterilized 3D-printed scaffolds at standardized densities (e.g., 50,000 cells/scaffold).
Viability Assessment:
- Employ live/dead staining at predetermined intervals (e.g., 1, 3, 7 days).
- Quantify viability using fluorescence microscopy and image analysis software.
- Alternatively, use metabolic assays (e.g., MTT, Alamar Blue) at 24-hour intervals [10].
Cell Morphology and Differentiation:
- Fix cells and perform immunocytochemistry for neural markers (e.g., β-III-tubulin, GFAP).
- Image using confocal microscopy to assess cell morphology and differentiation.
- Quantify expression levels using flow cytometry or Western blotting where appropriate.
In Vivo Biocompatibility Assessment Protocol
Objective: Evaluate host response to implanted biomaterials in animal models [10].
Animal Model and Implantation:
- Use appropriate immunocompetent animal models (e.g., rats, mice).
- Perform surgical implantation of sterilized scaffolds according to approved ethical protocols.
- Include appropriate controls (sham surgery, commercially available materials).
Histological Analysis:
- Euthanize animals at predetermined endpoints (e.g., 10 days, 30 days).
- Harvest implantation sites with surrounding tissue.
- Process for histology: fix in formalin, dehydrate, embed in paraffin, section.
- Stain with Hematoxylin & Eosin for general morphology and Masson's Trichrome for collagen deposition.
- Score inflammatory response semi-quantitatively based on immune cell infiltration [10].
Immunohistochemical Analysis:
- Perform staining for specific immune markers (e.g., CD68 for macrophages, CD3 for T-lymphocytes).
- Assess fibrous capsule thickness and composition.
- Evaluate tissue integration and vascularization at the implant-tissue interface.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Research Reagents for Biomaterials Translation
| Reagent/Category | Function | Specific Examples | Application Notes |
|---|---|---|---|
| Structural Polymers | Provide 3D scaffold architecture and mechanical support | PLA, PCL, Polyurethane (Filaflex), SEBS (Flexdym) [10] | Thermoplastics offer superior printability; synthetic polymers provide tunable mechanical properties [10] |
| Biofunctional Hydrogels | Mimic extracellular matrix; support cell adhesion and signaling | GelMA (gelatin methacrylate) [10] | Photocrosslinkable; offers biofunctionality with mechanical tunability; superior cell viability [10] |
| Crosslinking Agents | Enable hydrogel solidification and stability | LAP (lithium phenyl-2,4,6-trimethylbenzoylphosphinate) [10] | Photoinitiator for UV crosslinking of methacrylated polymers; critical for maintaining structural fidelity |
| Cell Culture Reagents | Support in vitro cell viability, growth, and differentiation | Neural stem cells, Schwann cells, appropriate culture media [10] | Primary cells provide most relevant response; standardized media formulations essential for reproducibility |
| Characterization Reagents | Enable assessment of material properties and biological responses | Live/Dead staining kits, antibodies for immunostaining, histological stains [10] | Critical for quantifying cell-material interactions; validate biocompatibility claims |
| Cyclopentadiene-quinone (2 | Cyclopentadiene-quinone (2, MF:C16H16O2, MW:240.30 g/mol | Chemical Reagent | Bench Chemicals |
| Apoptosis inducer 4 | Apoptosis Inducer 4|RUO | Apoptosis Inducer 4 is a potent compound with anticancer research applications. This product is for Research Use Only and not for human use. | Bench Chemicals |
Navigating the Translation Pathway: From Evidence to Clinical Product
The journey from laboratory research to clinical product requires careful navigation of regulatory, manufacturing, and validation hurdles. The following diagram illustrates this complex translation pathway.
Figure 3: The Biomaterials Translation Pathway. This roadmap highlights key stages from basic research to clinical adoption, emphasizing the critical role of evidence synthesis in de-risking translation [4] [6].
Advanced Translation Strategies
Adaptive Validation Frameworks
Recent innovations propose adaptive validation frameworks that align evidence requirements with an AI tool's risk profile, integrating real-world evidence for more efficient clinical translation [11]. As stated by researchers developing these frameworks, "Accuracy isn't enough—what matters is impact on patient outcomes" [11]. This approach is particularly relevant for data-driven biomaterial discovery enabled by machine learning.
Immune-Compatible Design Strategies
Considering the host immune response throughout therapeutic development is crucial for translational success. Design strategies include:
- Biomaterial Surface Modification: Modulating hydrophilicity to control protein adsorption (e.g., hydrophobic surfaces for albumin adsorption to block cell adhesion) [6].
- Mechanical Property Optimization: Matching tissue-specific stiffness (e.g., softer materials for neural tissues) to modulate immune activation [6].
- Controlled Degradation Profiles: Using synthetic biomaterials that hydrolyze into non-toxic byproducts or incorporating enzyme-cleavable crosslinks for environmentally-responsive degradation [6].
The biomaterials translation roadmap is evolving from traditional empirical approaches toward evidence-based and data-driven methodologies. By systematically applying evidence hierarchies, employing standardized experimental protocols, and leveraging emerging technologies like machine learning, researchers can accelerate the development of safer, more effective biomaterials while reducing costly translational failures. The comparative data and methodologies presented in this guide provide a framework for researchers to design robust development strategies that generate high-quality evidence, ultimately bridging the gap between promising laboratory results and clinically impactful medical products.
In the rigorous fields of biomaterials science and drug development, the ability to distinguish between strong and weak evidence is fundamental. The hierarchy of evidence is a core principle of evidence-based medicine (EBM), providing a structured framework for ranking research methodologies based on their reliability and potential for bias [12]. This pyramid-shaped hierarchy ensures that clinical guidelines and research directions are informed by the most trustworthy evidence available, with systematic reviews and meta-analyses at its apex, followed by randomized controlled trials, observational studies, and expert opinions [8] [12]. For researchers and scientists, understanding this hierarchy is not merely academic; it is a critical tool for designing robust studies, critically appraising published literature, and making informed decisions that ultimately impact patient care and product development.
The concept of a hierarchy of evidence originated in the mid-20th century, championed by figures like Archie Cochrane, who emphasized the necessity of systematic reviews of randomized controlled trials (RCTs) [12]. This approach has since evolved into a more nuanced system, formalized by organizations like the Centre for Evidence-Based Medicine, which classifies evidence into levels to guide practitioners and policymakers [9]. In biomaterials research, where the translation of a new material from the lab to a commercial medical product is complex and costly, employing a sound evidence-based approach is increasingly recognized as vital for validating scientific findings and prioritizing research resources [4].
The Evidence Pyramid: A Detailed Breakdown
The hierarchy of evidence is commonly visualized as a pyramid. This section provides a detailed comparison of each level, from the most to the least methodologically rigorous.
Level 1: Systematic Reviews and Meta-Analyses
At the top of the pyramid are systematic reviews and meta-analyses, which represent the highest quality of evidence [8] [12]. A systematic review is a type of literature review that uses a systematic and predefined process to identify, select, and critically appraise all available research on a specific, focused question [13]. A meta-analysis is a statistical component that quantitatively combines and synthesizes the results of the selected studies, thereby providing a more precise estimate of an intervention's effect [13].
The primary strength of this level is its comprehensive nature and its ability to minimize bias through a structured, transparent, and reproducible methodology [12]. By synthesizing findings from multiple studies, systematic reviews can resolve uncertainties and contradictions in the literature. However, it is crucial to note that the quality of a systematic review is entirely dependent on the quality of the individual studies it includes; a review of flawed or biased primary studies will produce similarly flawed conclusions [12].
Level 2: Randomized Controlled Trials (RCTs)
Just below systematic reviews are Randomized Controlled Trials (RCTs). In an RCT, participants are randomly allocated to either an intervention group or a control group. This random allocation is a powerful methodological feature that minimizes selection bias and helps ensure that the groups are comparable at the start of the trial [12]. This design allows researchers to establish causal relationships between an intervention and an outcome with high confidence.
RCTs are considered the gold standard for evaluating the efficacy of new therapies or interventions. Nevertheless, they have limitations: they can be resource-intensive and expensive, may raise ethical concerns in certain situations, and their highly controlled setting can sometimes limit the generalizability of their findings to real-world clinical practice [12].
Level 3: Observational Studies (Cohort and Case-Control Studies)
Occupying the middle of the pyramid are observational studies, which include cohort studies and case-control studies. Unlike RCTs, researchers in observational studies do not assign interventions; they instead observe and analyze the effect of an exposure or intervention as it occurs naturally.
- Cohort Studies: These follow a group of people (a cohort) over time to see how different exposures affect outcomes. They can be prospective (following groups forward in time) or retrospective (looking back at historical data) [12]. Prospective designs generally provide more reliable data.
- Case-Control Studies: These start with individuals who already have a disease (cases) and compare them to similar individuals without the disease (controls), looking back to identify differences in exposure [9].
While observational studies are invaluable for investigating questions where RCTs are not feasible (e.g., studying the harmful effects of smoking), they are more susceptible to confounding variables, which can obscure the true relationship between an exposure and an outcome [12].
Level 4: Case Series and Case Reports
Case series and case reports provide detailed descriptions of a single patient or a small group of patients with a similar condition or treatment. They are primarily hypothesis-generating and are particularly useful for reporting rare diseases, unexpected treatment effects, or novel surgical techniques [12].
Due to their small scale, lack of a control group, and inherent selection bias, they sit low on the evidence hierarchy. Their findings are not generalizable to the broader population, but they can serve as a critical first step in identifying new areas for more rigorous investigation [12].
Level 5: Expert Opinion and Anecdotal Evidence
At the base of the pyramid are expert opinions and anecdotal evidence. This level relies on the personal experience and judgment of authorities in the field rather than systematic research [8] [12]. While expert opinion can be insightful for addressing complex clinical questions where higher-level evidence is lacking, it is considered the least reliable form of evidence. It is highly subjective, prone to bias, and lacks the systematic methodology and controls necessary for scientific validation [12].
Table 1: Summary of Evidence Levels in Medical and Biomaterials Research
| Level | Study Type | Key Features | Strengths | Weaknesses |
|---|---|---|---|---|
| 1 | Systematic Review & Meta-Analysis | Synthesizes multiple high-quality studies using a predefined protocol [13] [12]. | Highest level of evidence, minimizes bias, provides comprehensive conclusions [12]. | Quality dependent on included studies; can be complex and time-consuming [12]. |
| 2 | Randomized Controlled Trial (RCT) | Participants randomly assigned to intervention or control groups [12]. | Establishes causation, minimizes selection bias [12]. | Can be costly, may lack generalizability, ethical constraints for some questions [12]. |
| 3 | Cohort Study | Observes groups with/without exposure over time [12]. | Suitable for studying etiology and incidence, can study multiple outcomes [12]. | Susceptible to confounding, time-consuming, not suitable for rare diseases [12]. |
| 3 | Case-Control Study | Compares subjects with a disease (cases) to those without (controls) [9]. | Efficient for rare diseases, requires fewer subjects, can study multiple exposures [12]. | Prone to recall and selection bias, cannot establish incidence or causality [12]. |
| 4 | Case Series / Report | Detailed report on one or a small group of patients [12]. | Identifies new diseases or adverse events, hypothesis-generating [12]. | No control group, susceptible to bias, low generalizability [12]. |
| 5 | Expert Opinion | Based on personal experience and judgment [8] [12]. | Useful when no research evidence exists, provides quick insights [12]. | Subjective, lacks systematic methodology, highly prone to bias [12]. |
The following diagram illustrates the standard evidence hierarchy and the foundational workflow for generating its highest level of evidence: the systematic review.
Experimental Protocols in Biomaterials Research
The principles of the evidence hierarchy are directly applicable to biomaterials research. The following examples illustrate how different study designs are implemented in this field.
Protocol for a Systematic Review
The systematic review titled "A Histological and Clinical Evaluation of Long-Term Outcomes of Bovine-Derived Xenografts" provides a clear template for this level of evidence [14].
- Research Question Formulation: The review used the PICO framework (Population, Intervention, Comparison, Outcome) to define its scope. The population was patients undergoing dental implant surgery requiring bone regeneration; the intervention was bovine-derived xenografts; the comparison was alternative grafting materials; and the outcomes were long-term clinical and histological results [14].
- Search Strategy: A comprehensive electronic search was performed across multiple databases (PubMed, Scopus, Web of Science) using a combination of keywords like "Bio-Oss," "bovine bone graft," "bone regeneration," and "long-term" connected by Boolean operators (AND, OR) [14].
- Screening & Selection: The process followed the PRISMA guidelines. After duplicate removal, studies were screened by title/abstract and then by full text against predefined inclusion and exclusion criteria [14].
- Data Extraction & Quality Assessment: Two independent reviewers performed data extraction. The quality of evidence was further assessed using the GRADE framework, which evaluates risk of bias, inconsistency, indirectness, imprecision, and publication bias [14].
Protocol for an In Vivo Animal Study
The systematic review on "Whitlockite as a next-generation biomaterial for bone regeneration" summarizes the protocols common to many of the animal studies it synthesized [15].
- Animal Models: The included studies utilized animal bone defect models in species such as rats, mice, and rabbits to test the efficacy of Whitlockite-based materials [15].
- Intervention: The biomaterial (Whitlockite in various forms like nanoparticles, granules, and composite scaffolds) was implanted into the bone defects [15].
- Comparison: The test groups were compared against control groups, often treated with conventional materials like hydroxyapatite (HA) or β-tricalcium phosphate (β-TCP) [15].
- Outcome Measures: Key outcomes included quantitative measures of bone regeneration, such as bone volume fraction (BV/TV) and bone mineral density (BMD), as well as histological analysis of bone quality and expression of osteogenic markers (e.g., ALP, OCN) [15].
- Quality Assessment: The reviewers used the SYRCLE Risk of Bias Tool, a specific tool for assessing bias in animal studies [15].
Table 2: Comparison of Biomaterials Study Designs and Outcomes
| Study Focus | Study Type | Level of Evidence | Primary Outcome Measures | Key Findings |
|---|---|---|---|---|
| Bovine-Derived Xenografts [14] | Systematic Review of Human Studies | Level 1 (if including RCTs) | Long-term graft integration, complication rates (inflammation, migration), implant survival [14]. | Favorable long-term outcomes and implant survival, but concerns about late-onset complications and limited biodegradability [14]. |
| Whitlockite Biomaterials [15] | Systematic Review of Animal Studies | Level 1 (for animal evidence) | Bone volume fraction (BV/TV), bone mineral density (BMD), osteogenic marker expression (ALP, OCN) [15]. | Whitlockite consistently outperformed traditional materials (HA, β-TCP), showing 2–6% greater BV/TV and BMD, with enhanced osteogenesis [15]. |
| Biodegradable Orthopaedic Implants [16] | Meta-Analysis & Statistical Review | Level 1 | Tensile strength, degradation rate, biocompatibility [16]. | Biodegradable materials showed sufficient strength for orthopaedic applications, with degradation rates in the pattern BME > NBME > BHE > BLE [16]. |
Conducting high-quality, evidence-based research requires a suite of methodological tools and resources. The table below details key solutions used across different stages of the research process.
Table 3: Research Reagent Solutions for Evidence-Based Biomaterials Research
| Tool / Resource | Category | Primary Function | Application in Research |
|---|---|---|---|
| PICO / PICOTTS [13] | Methodological Framework | Provides a structured approach to formulating a focused, answerable research question [13]. | Defines Population, Intervention, Comparison, Outcome, and optionally Time, Type of study, and Setting. Foundation of any systematic review [14]. |
| PubMed / MEDLINE [13] | Bibliographic Database | Provides access to a vast repository of life sciences and biomedical literature [13]. | A primary database for conducting comprehensive literature searches, often used with MeSH terms and Boolean operators [13] [14]. |
| Covidence / Rayyan [13] | Research Software | Streamlines the systematic review process by assisting with study screening, selection, and data extraction [13]. | Improves efficiency and accuracy during the systematic review workflow, enabling collaboration among reviewers [13]. |
| Cochrane Risk of Bias Tool [13] | Quality Assessment | A standardized tool for assessing the methodological quality and risk of bias in randomized controlled trials [13]. | Used in systematic reviews to evaluate the internal validity of included RCTs, informing the strength of the overall evidence [13]. |
| SYRCLE Risk of Bias Tool [15] | Quality Assessment | A tool specifically designed to assess the risk of bias in animal studies [15]. | Critical for ensuring the quality and reliability of pre-clinical evidence synthesized in systematic reviews of animal literature [15]. |
| GRADE Framework [14] | Quality Assessment | A systematic approach to rating the overall quality of a body of evidence and strength of recommendations [14]. | Used to evaluate evidence across domains (bias, inconsistency, etc.), allowing for transparent judgment of evidence certainty [14]. |
| R / RevMan [13] | Statistical Software | Specialized software for performing meta-analysis and generating statistical summaries (e.g., forest plots) [13]. | Used to compute pooled effect sizes, confidence intervals, and assess heterogeneity across studies in a meta-analysis [13]. |
The following diagram maps the logical progression of a research idea through different study designs, highlighting the iterative nature of evidence generation and the role of technological advancements.
Navigating the hierarchy of evidence is an essential skill for any professional engaged in biomaterials research or drug development. From the foundational observations of case reports to the definitive power of systematic reviews, each level of evidence plays a distinct and valuable role in the scientific ecosystem. A clear understanding of the relative strengths, weaknesses, and appropriate applications of each study design allows researchers to not only produce more robust and impactful science but also to critically evaluate the work of others. As the field continues to evolve with the integration of artificial intelligence, big data, and real-world evidence, the fundamental principles of this hierarchy will remain the bedrock of evidence-based practice, ensuring that advancements in biomaterials are built upon a foundation of rigorous, transparent, and reliable scientific evidence [12] [17].
The field of biomaterials has evolved from a materials-centric discipline to a multidisciplinary science that increasingly emphasizes biological interactions and clinical applications [18]. This rapid development has generated an enormous volume of research data, creating an urgent need for methodologies that can translate this data into validated scientific evidence [18]. The concept of evidence-based biomaterials research (EBBR) has emerged to address this need, applying systematic approaches to evaluate and synthesize research findings [18]. Central to this evaluation is understanding the distinction between primary and secondary information sources, as this distinction fundamentally impacts the reliability and level of evidence available for making scientific and clinical decisions in biomaterials development and application.
In biomaterials research, the origin of data plays a critical role in assessing its validity as scientific evidence. The classification into primary and secondary sources determines not only how the data is collected but also its potential limitations and appropriate applications.
Primary data refers to information collected directly by researchers specifically for the purpose of their study using ad hoc methods [19]. In biomaterials research, this typically includes:
- Data from original laboratory experiments investigating material synthesis, processing, structure, and properties [18]
- Biocompatibility and biosafety evaluations conducted per ISO 10993 standards [18]
- Pre-clinical animal studies designed to assess specific material performance and host responses [18]
- Objective diagnostic methods such as blood tests or imaging results specifically conducted for the research [19]
- Direct measurements of material properties and performance characteristics [18]
Secondary data comprises information that was originally collected for purposes other than the current research study and is repurposed for new investigations [19]. In biomaterials contexts, this includes:
- Prescription registers and medical records documenting device implantation outcomes [19]
- Computerized pharmacy records and insurance claims databases [19]
- Previously published research data that is reanalyzed for new insights [18]
- Clinical and administrative databases originally established for billing or other non-research purposes [19]
Table 1: Comparison of Primary and Secondary Data Sources in Biomaterials Research
| Characteristic | Primary Data | Secondary Data |
|---|---|---|
| Collection Purpose | Specifically for research questions | For non-research purposes (clinical, administrative) |
| Data Control | Researchers design collection methods | Researchers utilize existing data structures |
| Common Sources | Laboratory experiments, preclinical studies, clinical trials | Medical records, prescription databases, claims data |
| Key Advantages | Targeted to specific research questions, controlled conditions | Larger sample sizes, real-world clinical settings |
| Potential Limitations | Limited sample size, artificial experimental conditions | Missing variables, measurement bias, confounding factors |
Methodological Approaches for Evidence Generation
Experimental Protocols for Primary Data Collection
Generating robust primary evidence in biomaterials research requires standardized methodological approaches:
Material Synthesis and Characterization Protocol:
- Material Processing: Utilize sol-gel, 3D printing, or other fabrication techniques appropriate to the biomaterial class [18]
- Structure Analysis: Employ SEM, TEM, or XRD to characterize material microstructure [18]
- Property Assessment: Conduct mechanical testing, degradation studies, and surface analysis [18]
- Biological Evaluation: Perform in vitro cell culture studies to assess cytotoxicity and cellular responses [18]
Pre-clinical Evaluation Workflow:
- Animal Model Selection: Choose species-specific models (e.g., calvarial defect models for bone regeneration) [18]
- Implantation Procedure: Standardize surgical protocols and control groups [18]
- Outcome Assessment: Implement histomorphometry, micro-CT, and biomechanical testing at predetermined endpoints [18]
- Statistical Analysis: Apply appropriate power calculations and statistical methods to ensure reproducible results [18]
Systematic Review Methodology for Secondary Data Analysis
The evidence-based approach utilizes systematic reviews to synthesize existing research:
Protocol Development:
- Formulate specific research questions using PICO (Population, Intervention, Comparison, Outcome) framework [18]
- Establish explicit inclusion and exclusion criteria for studies [18]
- Define comprehensive search strategies across multiple databases [18]
Data Extraction and Synthesis:
- Develop standardized data extraction forms to capture key study characteristics and outcomes [18]
- Implement quality assessment of individual studies using validated tools [18]
- Conduct meta-analysis when appropriate to quantitatively synthesize results [18]
Comparative Analysis: Evidence Levels and Applications
The distinction between primary and secondary data sources carries significant implications for the level of evidence they provide in the biomaterials translation roadmap from basic research to commercialized medical products [18].
Table 2: Evidence Levels and Applications in Biomaterials Translation
| Research Stage | Primary Data Applications | Secondary Data Applications | Evidence Level |
|---|---|---|---|
| Basic Research | Hypothesis testing, mechanism exploration, structure-property relationships | Literature reviews, research trend analysis, knowledge gap identification | Foundational |
| Applied Research | Prototype development, in vitro performance testing, preliminary safety assessment | Systematic reviews, meta-analyses of existing studies, research synthesis | Preclinical |
| Product Development | Biocompatibility testing, animal studies, design verification and validation | Post-market surveillance data analysis, clinical registry reviews | Regulatory |
| Clinical Evaluation | Controlled clinical trials, specific performance metrics | Real-world evidence (RWE), clinical database analysis, outcomes research | Clinical Validation |
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Research Reagents and Materials for Biomaterials Evidence Generation
| Reagent/Material | Function in Research | Primary/Secondary Application |
|---|---|---|
| Sol-gel Bioactive Glasses | Dental and periodontal tissue regeneration studies [18] | Primary data generation through in vitro and in vivo testing |
| 3D Printed Scaffolds | Bone regeneration research in calvarial defect models [18] | Primary data on design parameters (material, porosity, pore size) |
| Cell Culture Assays | Biocompatibility assessment and host-response evaluation [18] | Primary data on cellular interactions with biomaterials |
| ISO 10993 Test Materials | Standardized biosafety evaluation for regulatory approval [18] | Primary data for design validation and verification |
| Medical Device Databases | Post-market surveillance and real-world performance analysis [18] | Secondary data analysis for safety and effectiveness monitoring |
| Clinical Registry Data | Long-term outcomes assessment of implanted devices [18] | Secondary data for real-world evidence generation |
| Propyl 2,4-dioxovalerate | Propyl 2,4-dioxovalerate, CAS:39526-01-7, MF:C8H12O4, MW:172.18 g/mol | Chemical Reagent |
| 4-Chloro-1-ethyl-piperidine | 4-Chloro-1-ethyl-piperidine, CAS:5382-26-3, MF:C7H14ClN, MW:147.64 g/mol | Chemical Reagent |
Analysis of Heterogeneity in Biomaterials Evidence
A critical challenge in evaluating biomaterials evidence lies in addressing heterogeneity between primary and secondary data sources. Current reporting standards in major medical journals demonstrate significant limitations in this regard:
- Only 34.8% of meta-analyses in high-impact journals report the source of data (primary vs. secondary) [19]
- Merely 13.0% include method of outcome assessment as a variable in heterogeneity analysis [19]
- Only one surveyed meta-analysis compared and discussed results considering different data sources [19]
This represents a substantial gap in evidence evaluation, as failing to account for data source heterogeneity may lead to misleading conclusions in meta-analyses of biomaterials effects [19]. The integration of primary and secondary data sources through systematic methodology is essential for comprehensive evidence assessment.
The distinction between primary and secondary information sources represents a fundamental consideration in evaluating scientific evidence for biomaterials. Primary data offers controlled, targeted insights specifically designed to answer research questions, while secondary data provides valuable real-world context and larger sample sizes. The emerging methodology of evidence-based biomaterials research emphasizes systematic approaches to synthesizing both types of evidence, with careful consideration of source heterogeneity [18]. As the field advances, explicit acknowledgment and methodological rigor in handling these distinct information sources will be crucial for generating reliable evidence to guide biomaterials development, regulatory approval, and clinical application, ultimately supporting the translation of innovative biomaterial technologies from concept to clinical practice [18].
The Critical Role of Evidence in Regulatory Approval and Clinical Translation
The journey of a biomaterial from a laboratory concept to a clinically approved medical product is governed by a rigorous, evidence-based pathway. For researchers, scientists, and drug development professionals, understanding the hierarchy of evidence and specific regulatory requirements is crucial for successful clinical translation. The global biomaterials market, projected to grow from USD 192.43 billion in 2025 to approximately USD 523.75 billion by 2034, underscores the critical importance of this field and the evidence that supports it [20]. This guide examines the levels of evidence different biomaterial research studies generate, objectively compares regulatory strategies, and details the experimental protocols required to demonstrate safety and efficacy, providing a framework for navigating the complex translation from bench to bedside.
Regulatory Frameworks and Evidence Requirements
FDA Pathways and Evidence Standards
The U.S. Food and Drug Administration (FDA) employs distinct regulatory pathways for medical devices and biomaterials, each with unique evidence requirements [21]:
- 510(k) Clearance: Requires demonstration of "substantial equivalence" to a legally marketed predicate device. Evidence typically includes comparative bench testing, preclinical data, and possibly limited clinical data.
- Premarket Approval (PMA): Mandatory for high-risk devices (Class III) with no predicate, requiring extensive scientific evidence, including robust clinical trial data.
- De Novo Classification: For novel, low-to-moderate risk devices with no predicate, requiring evidence of safety and effectiveness through analytical, preclinical, and clinical data as needed.
The FDA has increasingly incorporated Real-World Evidence (RWE) into regulatory decision-making. As shown in , RWE has supported various regulatory actions, including new approvals and label changes, across multiple medical product types [22].
Table 1: FDA Use of Real-World Evidence in Regulatory Decisions (Select Examples)
| Product | Data Source | Study Design | Regulatory Action | Role of RWE |
|---|---|---|---|---|
| Aurlumyn (Iloprost) [22] | Medical records | Retrospective cohort study | New drug approval (Feb 2024) | Confirmatory evidence |
| Vimpat (Lacosamide) [22] | PEDSnet data network | Retrospective cohort study | Labeling change (Apr 2023) | Safety data for new dosing |
| Actemra (Tocilizumab) [22] | National death records | Randomized controlled trial | Approval (Dec 2022) | Primary efficacy endpoint in an AWC* study |
| Prolia (Denosumab) [22] | Medicare claims data | Retrospective cohort study | Boxed Warning (Jan 2024) | Identified risk of severe hypocalcemia |
Note: AWC = Adequate and Well-Controlled
Accelerated Approval and Confirmatory Evidence
For serious conditions with unmet medical needs, the Accelerated Approval Pathway allows approval based on surrogate endpoints (e.g., laboratory measures or physical signs) that are "reasonably likely to predict clinical benefit" [23]. This pathway is critical for accelerating patient access to novel therapies, including advanced biomaterials. However, it mandates post-approval confirmatory trials to verify the anticipated clinical benefit. Recent FDA guidances emphasize that these confirmatory trials should be underway at the time of approval, with clearly defined milestones to ensure timely completion [23]. Failure to verify clinical benefit in confirmatory trials or to demonstrate safety can lead to product withdrawal from the market.
Hierarchy of Evidence in Biomaterials Research
The strength of conclusions drawn from biomaterials research depends fundamentally on the study design. The "hierarchy of evidence" ranks research designs based on their ability to minimize bias and establish causality [24].
Figure 1: The Hierarchy of Evidence for Biomaterials Research. Experimental designs like RCTs offer the highest internal validity for establishing cause-and-effect, while descriptive studies provide foundational knowledge but are more susceptible to bias [24].
Quantitative Research Designs and Their Evidence Level
Descriptive (Non-Experimental) Designs: These observe and describe patterns without attempting to modify the subject of study. They are foundational but occupy the lower levels of the evidence hierarchy due to limitations in establishing causality [24].
- Cross-Sectional Studies: Provide a "snapshot" in time, useful for determining prevalence. They are relatively inexpensive and convenient but cannot establish temporal relationships [24].
- Case-Control Studies: Begin with the outcome (e.g., implant failure) and look backward for exposures or risk factors. They are efficient for studying rare outcomes but prone to selection and recall bias [24].
- Cohort Studies: Follow groups of individuals (cohorts) over time, either prospectively (forward in time) or retrospectively (using historical data). They can establish temporal sequence and are used to study multiple outcomes from a single exposure [24].
Experimental Designs: These involve direct manipulation of an independent variable (e.g., a new biomaterial) under controlled conditions.
- Randomized Controlled Trials (RCTs): The "gold standard" for establishing efficacy. Participants are randomly allocated to intervention or control groups, minimizing selection bias and confounding. RCTs provide the highest level of evidence for a single study [24].
- Quasi-Experimental Designs: Implement an intervention but lack full control, such as random assignment. They are used when RCTs are not feasible due to ethical or practical constraints but are more vulnerable to threats in internal validity [24].
Comparative Analysis of Biomaterials and Regulatory Strategies
The choice of biomaterial and corresponding regulatory strategy depends on the application's specific requirements, including mechanical properties, biodegradability, and bioactivity.
Table 2: Comparison of Major Biomaterial Classes and Their Applications
| Biomaterial Class | Key Characteristics | Common Applications | Regulatory Considerations |
|---|---|---|---|
| Polymers [20] [25] | Versatile, tunable degradation, can be natural or synthetic. | Tissue engineering scaffolds, drug delivery systems, resorbable sutures, hydrogels. | Often Class II devices (510(k)); biocompatibility testing per ISO 10993 is critical. |
| Metals [20] | High strength, fatigue resistance, load-bearing capacity. | Orthopedic implants (joint replacements, plates), dental implants, stents. | Often Class III devices (PMA); require extensive mechanical and fatigue testing. |
| Ceramics [20] [26] | High compressive strength, bioactive, inert. | Dental implants, bone graft substitutes, joint replacements. | Bioceramics for bone contact often require clinical data demonstrating osteointegration. |
| Natural Biomaterials [20] | High biocompatibility, inherent bioactivity, biodegradable. | Wound healing, soft tissue regeneration, drug delivery. | Complex manufacturing and sourcing require rigorous validation; immunogenicity risk assessment. |
Table 3: Analysis of Regulatory Pathways for Medical Devices
| Regulatory Pathway | Risk Classification | Key Evidence Requirements | Typical Timeline | Strengths | Weaknesses |
|---|---|---|---|---|---|
| 510(k) Clearance [21] | Class I/II (Low-Moderate) | Substantial equivalence to a predicate; bench and animal testing. | ~90 days | Lower cost, faster time-to-market. | Requires a suitable predicate; not suitable for truly novel technologies. |
| De Novo Request [21] | Class I/II (Novel, Low-Moderate) | Evidence of safety and effectiveness; risk-benefit analysis; clinical data may be needed. | 150-200 days | Creates a new regulatory classification for novel devices. | More resource-intensive than 510(k); requires demonstration of low-to-moderate risk. |
| Premarket Approval (PMA) [21] | Class III (High Risk) | Extensive scientific evidence; typically requires clinical trials; manufacturing info. | 6 months - 1 year | Suitable for novel, high-risk, life-sustaining devices. | Highest cost and longest timeline; requires intensive clinical data. |
Experimental Protocols and Methodologies for Biomaterials
Generating robust evidence for regulatory submissions requires adherence to standardized experimental protocols. Below are detailed methodologies for key areas of biomaterial testing.
Preclinical Biocompatibility and Safety Testing
Before human studies, biomaterials must undergo rigorous preclinical safety assessment, aligned with international standards like ISO 10993 ("Biological evaluation of medical devices") [21].
Protocol: Cytotoxicity Testing (ISO 10993-5)
- Objective: To determine if the biomaterial leaches toxic substances that cause cell death or damage.
- Methodology:
- Sample Preparation: Extract the biomaterial using both polar and non-polar solvents under standardized conditions (e.g., 37°C for 24 hours).
- Cell Culture: Use established mammalian cell lines (e.g., L-929 mouse fibroblast cells) cultured in appropriate media.
- Exposure: Apply the extract to the cell monolayers, with control groups receiving only the extraction vehicles.
- Incubation and Analysis: Incubate for 24-72 hours. Assess cell viability using quantitative methods like the MTT assay, which measures mitochondrial activity, or by qualitative microscopic examination for morphological changes.
- Data Interpretation: A reduction in cell viability by more than 30% is generally considered a sign of potential cytotoxicity.
Protocol: Sensitization Testing (ISO 10993-10)
- Objective: To evaluate the potential for the biomaterial to cause an allergic contact dermatitis response.
- Methodology (Murine Local Lymph Node Assay - LLNA):
- Animal Model: Use female mice (e.g., CBA/Ca or BALB/c strains).
- Application: Apply the biomaterial extract or control substances to the dorsum of both ears daily for three consecutive days.
- Pulsing and Measurement: On day five, inject the mice with radioactive thymidine. Five hours later, drain the auricular lymph nodes and measure the incorporation of radioactivity using a beta-scintillation counter.
- Data Interpretation: A stimulation index (SI) of three or greater compared to the vehicle control indicates a sensitizing potential.
Performance and Functional Testing
Protocol: Mechanical Testing for Orthopedic Implants
- Objective: To verify that an implant can withstand physiological loads without failure.
- Methodology:
- Sample Preparation: Manufacture test samples to final implant specifications.
- Testing Equipment: Use a servo-hydraulic or electromechanical test frame with environmental control (e.g., in saline at 37°C).
- Static Testing: Perform tensile, compressive, or bending tests to determine yield strength, ultimate tensile strength, and modulus of elasticity.
- Dynamic Testing: Conduct fatigue testing by applying cyclic loads representative of in vivo conditions (e.g., up to 10 million cycles for a hip stem) to determine the implant's endurance limit.
- Data Interpretation: Compare results against minimum performance standards (e.g., ASTM F382 for bone plates) or predicate devices.
Protocol: In Vivo Efficacy in an Animal Model
- Objective: To evaluate the functional performance and tissue integration of a biomaterial in a living organism.
- Methodology (Example: Critical-Sized Bone Defect Model):
- Animal Model: Use a validated species (e.g., rat, rabbit, or sheep).
- Surgical Procedure: Create a standardized defect in a long bone (e.g., femur) that will not heal spontaneously.
- Implantation: Implant the test biomaterial into the defect. Control groups may receive a standard of care (e.g., autograft) or be left untreated.
- Monitoring and Analysis: After a pre-defined period (e.g., 8-12 weeks), euthanize the animals and analyze the defect site using micro-CT for 3D bone formation, histology for tissue morphology and integration, and biomechanical testing for strength.
Visualization of the Clinical Translation Workflow
The path from concept to clinic is a multi-stage process with iterative feedback loops, where evidence generation is central to each decision point [27].
Figure 2: The Biomaterial Clinical Translation Pathway. This workflow illustrates the staged evidence generation required for regulatory approval, from initial concept to post-market monitoring, highlighting the iterative feedback from post-market data to future designs [21] [27].
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful biomaterials research relies on a suite of specialized reagents and analytical tools to characterize properties and assess biological performance.
Table 4: Key Research Reagent Solutions for Biomaterials Development
| Reagent/Material | Function and Application | Key Characteristics |
|---|---|---|
| AlamarBlue / MTT Reagents [25] | Colorimetric or fluorometric assays to quantify cell viability and proliferation in cytotoxicity and biocompatibility testing. | Provide a quantitative measure of metabolic activity; high sensitivity. |
| Primary Cells (e.g., Osteoblasts, Chondrocytes) [27] | Used in in vitro models to assess cell-material interactions in a tissue-specific context. | More physiologically relevant than cell lines; require specific culture conditions. |
| ELISA Kits | Measure specific protein secretion (e.g., inflammatory cytokines, osteogenic markers) to evaluate the host immune response or bioactivity of a material. | Highly specific and quantitative; essential for evaluating molecular-level responses. |
| Scanning Electron Microscopy (SEM) Reagents (e.g., glutaraldehyde, gold sputter coater) | Prepare samples for imaging the surface topography and microstructure of biomaterials, as well as cell adhesion and morphology. | Provides high-resolution, detailed images at the micro- and nano-scale. |
| PCR Assays (qRT-PCR) | Quantify gene expression levels to understand how a biomaterial influences cellular differentiation (e.g., towards osteogenic lineage) or inflammatory pathways. | Highly sensitive; allows for mechanistic studies of material-cell interactions. |
| ISO 10993 Biocompatibility Test Matrix [21] | A standardized suite of tests (e.g., for sensitization, irritation, systemic toxicity) required for regulatory submission. | Internationally recognized framework for safety assessment. |
| Spikenard extract | Spikenard Extract | |
| p-O-Methyl-isoproterenol | p-O-Methyl-Isoproterenol|Supplier | p-O-Methyl-Isoproterenol (CAS 3413-49-8), a key metabolite of Isoproterenol. For Research Use Only. Not for human or veterinary diagnostic or therapeutic use. |
The clinical translation of biomaterials is a complex but structured process anchored in the generation of robust, hierarchical evidence. From basic biocompatibility testing to advanced RCTs and post-market surveillance, each stage of development builds upon the previous one to establish a comprehensive safety and efficacy profile. The regulatory landscape is dynamic, with pathways like Accelerated Approval and the growing use of RWE offering opportunities to accelerate patient access, while simultaneously placing greater emphasis on post-market validation. For researchers and developers, success hinges on a strategic understanding of these evidence requirements, early engagement with regulatory bodies, and the meticulous execution of standardized experimental protocols. As the field advances with AI-driven material design and personalized biomaterials, the foundational principles of evidence-based research will remain more critical than ever in bringing safe and effective biomaterial innovations to the clinic.
Generating Robust Evidence: Methodologies and Real-World Applications in Biomaterials
In the rapidly advancing field of biomaterials research, where new materials and applications emerge constantly, the ability to distinguish truly effective innovations from merely promising ideas is paramount. Systematic reviews and meta-analyses occupy the highest position in the evidence hierarchy, providing the most reliable foundation for scientific and clinical decision-making [28] [29]. These methodologies offer a structured, transparent, and reproducible framework for synthesizing research findings, effectively distinguishing signal from noise across sometimes contradictory primary studies.
The growing complexity of biomaterials science, encompassing everything from biodegradable alloys to tissue engineering scaffolds, has generated "significant number of studies and publications as well as tremendous amount of research data" [4]. This abundance creates an urgent need for evidence-based biomaterials research (EBBR), an approach that adapts the rigorous principles of evidence-based medicine to translate raw research data into validated scientific evidence through systematic methodology [4]. This article provides a comprehensive comparison of these two gold-standard evidence synthesis methods, detailing their protocols, applications, and implementation within the specific context of biomaterials research.
Core Concepts and Definitions
Systematic Reviews
A systematic review is a rigorous, structured research method that aims to identify, evaluate, and summarize all available evidence on a specific research question using a predefined, documented protocol [28] [13]. Unlike traditional narrative reviews, systematic reviews employ explicit, systematic methods to minimize bias and provide a comprehensive overview of the evidence landscape [28]. The primary purpose is to "gather all existing evidence on a particular topic, evaluate the quality of that evidence, create a transparent summary that answers a specific research question, identify gaps in current knowledge, and inform evidence-based practice and policy decisions" [28].
Meta-Analyses
A meta-analysis is a statistical procedure that combines the numerical results from multiple similar studies to calculate an overall effect size [28] [13]. Think of it as a "study of studies" that uses statistical methods to find the signal amid the noise of individual research findings [28]. While a systematic review asks, "What does all the evidence say?", a meta-analysis asks more specifically, "What is the mathematical average effect across all studies, and how confident can we be in this number?" [28]. A meta-analysis typically builds upon a systematic review foundation but requires additional steps including statistical pooling and weighting of studies based on their precision [28].
Key Differences Between Systematic Reviews and Meta-Analyses
While systematic reviews and meta-analyses are often mentioned together, they serve distinct purposes and employ different methodologies. The table below summarizes their key differences:
Table 1: Fundamental Differences Between Systematic Reviews and Meta-Analyses
| Feature | Systematic Review | Meta-Analysis |
|---|---|---|
| Definition | Comprehensive review that identifies, evaluates, and synthesizes all available evidence on a specific question [28] | Statistical technique that combines results from multiple similar studies to calculate an overall effect [28] |
| Primary Purpose | To gather and critically appraise all relevant research [28] | To provide a precise mathematical estimate of effect [28] |
| Nature | Primarily qualitative synthesis [28] | Primarily quantitative analysis [28] |
| Study Types Included | Can include diverse study designs [28] | Requires studies with compatible numerical data [28] |
| Analysis Method | Narrative synthesis, thematic analysis [28] | Fixed or random effects models, meta-regression [28] |
| Output Format | Text summary, evidence tables, narrative synthesis [28] | Forest plots, pooled effect sizes, confidence intervals [28] |
| Typical Conclusion | "The evidence suggests that..." [28] | "The pooled effect size is X (95% CI: Y-Z)" [28] |
A key conceptual relationship is that a meta-analysis typically builds upon a systematic review but requires the additional step of statistical pooling. All meta-analyses should be based on a systematic review, but not all systematic reviews include a meta-analysis [28] [13]. A meta-analysis is only possible when the included studies report compatible statistical outcomes that can be mathematically combined [28].
Experimental Protocols and Methodological Standards
Conducting high-quality evidence synthesis requires adherence to strict methodological standards. The following workflow outlines the key stages, which incorporate both systematic review and potential meta-analysis components:
Evidence Synthesis Workflow
Protocol Development and Registration
The process begins with developing and registering a detailed protocol, which serves as a research plan that minimizes bias and enhances transparency [29] [30]. The protocol should specify the research question, inclusion/exclusion criteria, search strategy, data extraction methods, and planned synthesis approaches [29]. Registration on platforms like PROSPERO protects against duplicate publication and reduces publication bias [30].
Formulating the Research Question
A well-structured research question is fundamental. In biomaterials research, frameworks like PICO (Population, Intervention, Comparator, Outcome) or its extended versions are commonly used [13] [30]. For example, in evaluating a new biodegradable material for orthopaedic implants:
- Population: Patients with long bone fractures requiring internal fixation
- Intervention: Biodegradable low-entropy alloy implants
- Comparator: Traditional non-biodegradable medium-entropy alloy implants
- Outcome: Fracture healing rate, implant degradation rate, adverse events [16]
Comprehensive Literature Search
A systematic search across multiple databases is essential to capture all relevant evidence. For biomaterials topics, this typically includes PubMed/MEDLINE, Embase, Web of Science, and specialized databases like Cochrane Library [13] [31]. The search strategy should combine controlled vocabulary (e.g., MeSH terms) with free-text keywords and include "gray literature" to reduce publication bias [13] [31]. Tools like EndNote, Zotero, or Mendeley help manage references and remove duplicates [13].
Study Selection and Data Extraction
Study selection involves screening titles, abstracts, and full texts against predefined inclusion/exclusion criteria, typically performed by at least two independent reviewers to minimize error [13] [32]. Data extraction uses standardized forms to collect information on study characteristics, participants, interventions, comparisons, outcomes, and methodology [13]. Recent advances show AI tools like GPT models can significantly improve screening efficiency, with one study showing GPT models achieving precision of 0.51 compared to 0.21 for Abstrackr, though Abstrackr showed slightly higher recall (0.74 vs. 0.79) [32].
Quality Assessment and Risk of Bias
Assessing the methodological quality and risk of bias of included studies is crucial for interpreting findings. Various validated tools exist for different study designs, including Cochrane Risk of Bias Tool for randomized trials and Newcastle-Ottawa Scale for observational studies [13] [33]. In a recent meta-analysis of AI diagnostic performance, 76% of included studies were rated at high risk of bias, primarily due to small test sets and unknown training data for AI models [33].
Data Synthesis Approaches
Narrative Synthesis
When studies are too methodologically diverse or report incompatible outcomes, a narrative synthesis provides a structured qualitative summary. This involves organizing findings thematically, tabulating study characteristics, and describing patterns across studies [28] [13].
Meta-Analysis
When studies report compatible quantitative data, meta-analysis provides a statistical method for pooling results. The key steps include:
- Calculating Effect Sizes: Determining appropriate effect measures (odds ratios, risk ratios, mean differences, etc.) for each study [13]
- Weighting Studies: Assigning weights to studies, typically based on their precision (inverse variance), giving more influence to larger studies [29]
- Choosing a Model: Selecting between fixed-effect (assumes one true effect size) or random-effects (allows for variation in true effects) models based on heterogeneity assessment [29]
- Statistical Analysis: Combining effect sizes using weighted average methods, Mantel-Haenszel odds ratios, or Peto method for rare events [29]
Table 2: Performance Comparison of AI Tools in Systematic Review Screening
| Performance Metric | Abstrackr | GPT Models | Statistical Significance |
|---|---|---|---|
| Recall | 0.740 (95% CI: 0.725, 0.755) | 0.791 (95% CI: 0.781, 0.801) | p < 0.001 [32] |
| Precision | 0.119 (95% CI: 0.114, 0.124) | 0.465 (95% CI: 0.456, 0.474) | p < 0.001 [32] |
| Specificity | 0.780 (95% CI: 0.776, 0.784) | 0.973 (95% CI: 0.968, 0.977) | p < 0.001 [32] |
| F1 Score | 0.206 (95% CI: 0.199, 0.213) | 0.591 (95% CI: 0.581, 0.601) | p < 0.001 [32] |
Applications in Biomaterials Research: Case Studies
Case Study 1: Evaluating Biodegradable versus Non-Biodegradable Orthopaedic Implants
A recent systematic review with meta-analysis compared biodegradable and non-biodegradable materials for orthopaedic applications [16]. The analysis revealed distinct patterns in material properties: for tensile strength, NBME (non-biodegradable medium entropy) > BHE (biodegradable high entropy) > BME (biodegradable medium entropy) > BLE (biodegradable low entropy) alloys, while degradation rates followed the pattern BME > NBME > BHE > BLE [16]. This quantitative synthesis demonstrated that biodegradable materials (BLE and BME) offer viable alternatives to traditional non-biodegradable materials in orthopaedic applications, with the advantage of eliminating the need for removal surgeries [16].
Case Study 2: Diagnostic Performance of Generative AI in Medicine
A comprehensive meta-analysis of 83 studies compared the diagnostic performance of generative AI models against physicians [33]. The overall accuracy for generative AI models was 52.1% (95% CI: 47.0-57.1%), with no significant performance difference between AI models and physicians overall (p = 0.10) or non-expert physicians (p = 0.93) [33]. However, AI models performed significantly worse than expert physicians (difference in accuracy: 15.8%, p = 0.007) [33]. This synthesis provided crucial evidence for understanding AI's current capabilities and limitations in medical diagnostics.
Table 3: Essential Tools and Resources for Conducting Systematic Reviews and Meta-Analyses
| Tool Category | Specific Tools | Function and Application |
|---|---|---|
| Protocol Development | PROSPERO, Cochrane Template | Protocol registration and development [29] |
| Literature Search | PubMed, Embase, Web of Science, Cochrane Library | Comprehensive literature identification [13] |
| Reference Management | EndNote, Zotero, Mendeley | Storing references and removing duplicates [13] |
| Study Screening | Rayyan, Covidence, Abstrackr | Managing title/abstract and full-text screening [13] [32] |
| Data Extraction | Covidence, SRDR, DistillerSR | Systematic data extraction using standardized forms [13] |
| Quality Assessment | Cochrane RoB Tool, Newcastle-Ottawa Scale | Assessing methodological quality and risk of bias [13] |
| Statistical Analysis | R, RevMan, Stata | Conducting meta-analyses and creating forest plots [13] |
| Reporting | PRISMA Checklist | Ensuring transparent and complete reporting [13] |
Current Challenges and Methodological Limitations
Despite their strengths, systematic reviews and meta-analyses face several challenges. Their quality is entirely dependent on the available primary studies—flawed or biased primary studies will lead to unreliable syntheses ("garbage in, garbage out") [28] [29]. Publication bias remains a significant concern, as studies with significant or positive results are more likely to be published, potentially skewing overall conclusions [28] [29]. Statistical heterogeneity—when studies show widely varying effect sizes—can complicate interpretation and pooling of results [29]. The process is also notably resource-intensive, typically requiring 6-18 months and a team of researchers rather than a single individual [28].
The field is evolving with emerging methodologies aimed at addressing these challenges. Automation tools and artificial intelligence are increasingly being applied to reduce the manual burden of screening and data extraction [34] [32]. There is growing emphasis on open science practices, including data sharing and protocol registration, to enhance transparency and reproducibility [31] [30]. New approaches for synthesizing preclinical evidence are being developed to address the unique challenges of basic science research [31].
Systematic reviews and meta-analyses represent the gold standard for evidence synthesis in biomaterials research and beyond. While systematic reviews provide comprehensive, qualitative summaries of all available evidence on a specific question, meta-analyses offer quantitative precision through statistical pooling of compatible studies. These methodologies are particularly valuable in the complex field of biomaterials research, where they can help resolve contradictions across individual studies, provide precise estimates of material performance and biocompatibility, and identify gaps in the current evidence base to guide future research priorities. When conducted with rigorous adherence to methodological standards and transparent reporting, these powerful synthesis methods provide the most reliable evidence foundation for advancing biomaterials science and translating innovations into clinical practice.
Applying Evidence-Based Medicine (EBM) Principles to Biomaterials Evaluation
The rapid development of biomaterials science has generated vast amounts of research data, creating a critical need for methodologies that can translate this data into validated scientific evidence [4]. Evidence-Based Biomaterials Research (EBBR) addresses this need by applying evidence-based approaches, notably systematic reviews and meta-analyses, to evaluate scientific questions in biomaterials [18]. Originating from evidence-based medicine, this approach provides a structured framework for assessing the quality and reliability of evidence in biomaterials development [5].
The fundamental goal of EBBR is to enable more efficient translation of biomaterial technologies from basic research to commercialized medical products by generating robust scientific evidence for evaluating safety and performance [18]. This is particularly crucial given the multidisciplinary nature of biomaterials, which involves diverse fields from materials science to clinical medicine [4].
Levels of Evidence in Biomaterials Research
The evidence-based approach establishes a hierarchy of evidence quality, with systematic reviews of randomized controlled trials at the highest level, followed by individual randomized controlled trials, cohort studies, case-control studies, case series, and anecdotal case reports at the lowest level [4]. This hierarchy provides researchers with a structured framework for evaluating the reliability of different types of biomaterials studies.
Table 1: Evidence Levels in Biomaterials Research
| Evidence Level | Study Type | Key Characteristics | Typical Applications in Biomaterials |
|---|---|---|---|
| Level I | Systematic Reviews & Meta-Analyses | Comprehensive synthesis of multiple studies; highest evidence quality | Establishing overall efficacy of biomaterial classes; guiding clinical translation |
| Level II | Individual Randomized Controlled Trials | Experimental designs with random assignment; control groups | Comparing new biomaterials against standard treatments in clinical settings |
| Level III | Cohort Studies | Observational studies following groups over time | Evaluating long-term performance and safety of implanted biomaterials |
| Level IV | Case-Control Studies | Comparing cases with outcomes to controls without | Investigating causes of rare biomaterial-related complications |
| Level V | Case Series & Reports | Documentation of individual cases or small series | Reporting novel applications or unexpected biomaterial behaviors |
Comparative Evaluation of Biomaterial Performance
Analysis of Immune Response Across Biomaterial Types
Recent systematic reviews have quantified how different biomaterial classes elicit distinct biological responses, particularly regarding immune activation and tissue integration [35]. These analyses provide crucial evidence for material selection based on specific clinical applications.
Table 2: Comparative Immune Responses to Biomaterial Classes
| Biomaterial Class | Acute Inflammatory Response | Macrophage Polarization | Fibrous Encapsulation | Tissue Integration Quality | Key Supporting Evidence |
|---|---|---|---|---|---|
| Bioactive Ceramics | Moderate to Low | Predominantly M2 (pro-regenerative) | Minimal | Excellent | Systematic review of 25 in vivo studies showing enhanced regeneration [35] |
| Polymers (PCL) with modified surfaces | Low | M2-dominated response | Significantly reduced | Very Good | Study demonstrating increased VEGF, reduced pro-inflammatory chemokines [35] |
| Acellular Dermal Matrices (Human) | Mild, transient | Balanced M1/M2 transition | Minimal | Excellent | Comparative studies showing effective tissue integration [35] |
| Biological Meshes (Porcine) | Intense cellular infiltration | M1-dominated (inflammatory) | Significant | Variable | Evidence of antibody production and substantial inflammation [35] |
| Metallic Biomaterials | Variable | Dependent on corrosion products | Common with non-bioactive metals | Moderate to Good | Multiple cohort studies showing long-term stability issues with certain alloys |
Characterization Methods for Biomaterial Evaluation
Advanced characterization technologies provide critical data for evidence-based evaluation of biomaterials. Multi-Parametric Surface Plasmon Resonance (MP-SPR) and Quartz Crystal Microbalance with Dissipation (QCMD) monitoring offer complementary approaches for analyzing biomaterial interactions [36].
Table 3: Experimental Methods for Biomaterial Characterization
| Method | Key Measured Parameters | Evidence Level Generated | Application Examples | Technical Requirements |
|---|---|---|---|---|
| MP-SPR | Thickness, refractive index, interaction kinetics, surface coverage | Level II (controlled experiments) | Drug release monitoring, protein adsorption studies, layer-by-layer deposition | Specialized sensor slides, flow control systems, optical measurement capabilities |
| QCMD | Mass changes, viscoelastic properties, binding affinities, layer thickness | Level II (controlled experiments) | Biomaterial degradation tracking, protein conformation changes, hydration studies | Electrochemical compatibility, temperature control, solvent-resistant sensors |
| In Vivo Animal Models | Host response, tissue integration, immune activation, degradation | Level III (cohort studies) | Biocompatibility assessment, tissue regeneration evaluation, long-term implantation studies | Animal facility, histological processing, IHC staining capabilities |
| Systematic Review | Synthesis of multiple study findings, meta-analysis | Level I (highest evidence) | Evaluating overall biomaterial class performance, clinical translation guidance | Comprehensive literature search, predefined inclusion criteria, statistical synthesis capability |
Detailed Experimental Protocols for Biomaterial Assessment
Protocol 1: Systematic Review Methodology for Biomaterials
The evidence-based approach to biomaterials relies on systematic reviews conducted according to PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines [35]:
Protocol Development and Registration: Predefine research questions using PICO framework (Population, Intervention, Comparators, Outcomes). Register protocol with Open Science Framework or similar registry [35].
Comprehensive Search Strategy: Execute systematic searches across multiple databases (VHL, PubMed, SCOPUS, EMBASE, Web of Science). Combine search terms using Boolean operators with controlled vocabularies (MeSH, DeCS, Emtree) [35].
Study Selection Process: Implement dual-phase screening with at least two independent reviewers. Initial title/abstract screening followed by full-text evaluation using Rayyan or similar tools [35].
Data Extraction and Quality Assessment: Extract predefined data elements into structured tables. Assess study quality and risk of bias using appropriate tools for different study designs [35].
Data Synthesis and Analysis: Perform qualitative synthesis of findings. Conduct meta-analysis where appropriate with statistical analysis of combined data [35].
Protocol 2: Real-Time Biomaterial-Protein Interaction Analysis
MP-SPR methodology provides quantitative data on biomaterial interactions with biological components [36]:
Sensor Surface Preparation: Functionalize sensor slides with biomaterial coatings using dip-coating, spin-coating, or layer-by-layer deposition. Characterize baseline thickness and refractive index [36].
Experimental Parameter Setup: Establish physiological conditions (pH 7.4, 37°C, flow rate 50-100 μL/min). Prepare protein solutions at physiological concentrations (e.g., serum albumin at 40 mg/mL) [36].
Association Phase Measurement: Inject protein solution over biomaterial surface while continuously monitoring SPR angle shift. Maintain flow for sufficient time to reach binding saturation (typically 15-30 minutes) [36].
Dissociation Phase Monitoring: Switch to buffer flow to monitor dissociation of loosely bound proteins. Continue monitoring until stable baseline is achieved [36].
Data Analysis: Calculate binding kinetics (association rate ka, dissociation rate kd, equilibrium constant K_D) using appropriate binding models. Determine surface coverage and mass accumulation [36].
Visualizing the Evidence-Based Biomaterials Evaluation Workflow
Evidence Evaluation Workflow
Biomaterial Immune Response Pathways
Immune Response Pathways
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 4: Key Research Reagents for Biomaterial Evaluation
| Reagent/Material | Function | Specific Application Examples | Evidence Generation Capability |
|---|---|---|---|
| MP-SPR Sensor Slides | Enable real-time label-free biomolecular interaction analysis | Protein adsorption studies, drug release kinetics, layer-by-layer deposition monitoring | Level II evidence through quantitative interaction data [36] |
| QCMD Sensors with Electrochemical Kits | Provide viscoelastic characterization of soft biomaterials | Hydration studies, degradation monitoring, mechanical properties assessment | Level II evidence through mass and dissipation measurements [36] |
| Acellular Dermal Matrices | Serve as biological scaffold reference materials | Comparative studies of host response, tissue integration models | Level III evidence through in vivo evaluation [35] |
| Polycaprolactone (PCL) Scaffolds | Provide synthetic polymer platform for modification | Surface modification studies, macrophage polarization experiments | Level II evidence through controlled in vitro and in vivo studies [35] |
| Bioactive Glass Compositions | Enable bone regeneration studies | Dental and periodontal tissue regeneration models, osseointegration assessment | Level I evidence when included in systematic reviews [18] |
| Standardized Protein Solutions | Facilitate consistent protein adsorption studies | Serum protein adsorption, fibronectin binding studies | Level II evidence through reproducible binding assays [36] |
| 2-Fluoroethyl fluoroacetate | 2-Fluoroethyl fluoroacetate, CAS:459-99-4, MF:C4H6F2O2, MW:124.09 g/mol | Chemical Reagent | Bench Chemicals |
| Sulindac methyl derivative | Sulindac Methyl Derivative|Research Compound | Explore sulindac methyl derivatives for cancer, neuro, and oxidative stress research. This product is for Research Use Only (RUO). Not for human or veterinary use. | Bench Chemicals |
The application of EBM principles to biomaterials evaluation represents a paradigm shift from unstructured empirical approaches to development strategies entrenched in comprehensive data analysis [17]. Evidence-Based Biomaterials Research provides a structured framework for translating extensive research data into validated scientific evidence, ultimately accelerating the development of safer and more effective biomaterial-based medical products [4] [18]. As the field continues to evolve, closer integration of systematic review methodologies with emerging technologies like machine learning and high-throughput characterization will further enhance our ability to generate robust evidence for clinical translation [17].
Bioactive glasses (BGs) are a class of synthetic silica-based biomaterials that bond to living bone and stimulate new bone growth. This guide objectively compares the bone regeneration performance of bioactive glasses against traditional bone grafts and other alternatives. Key differentiators of bioactive glasses include their osteoconductivity (providing a scaffold for bone growth), osteoinductivity (stimulating progenitor cells to form bone), and inherent antimicrobial properties [37]. Beyond being a simple filler, bioactive glasses actively participate in the regeneration process through the controlled release of therapeutic ions such as silicon, calcium, and phosphorus, which create an osteogenic microenvironment [38].
The level of evidence supporting their use varies, from robust in vitro data and consistent positive results in animal studies to a growing number of promising clinical studies and meta-analyses. A 2025 meta-analysis concluded that bioactive glass leads to greater bone volume retention six months after surgery compared to autografts and results in less graft resorption [38]. Furthermore, systematic reviews have found bioactive glass to be non-inferior to established treatments like antibiotic-loaded calcium sulfate for managing bone infections [37]. This guide synthesizes experimental data and protocols to provide researchers and clinicians with a clear, evidence-based comparison.
Performance Comparison: Bioactive Glass vs. Alternatives
The following tables summarize key performance metrics for bioactive glasses compared to other common bone graft materials, based on pre-clinical and clinical data.
Table 1: Quantitative Performance Metrics from Pre-clinical and Clinical Studies
| Performance Metric | Bioactive Glass (BG) | Autogenous Bone Graft (Autograft) | Allograft | Calcium Phosphate Ceramics |
|---|---|---|---|---|
| Bone Volume Retention (6 months) | SMD = 0.796 (CI = 0.445–1.147) [38] | Baseline (Reference) | Variable, often lower than autograft | Generally lower resorption than BG [38] |
| Graft Resorption Rate | Significantly lower than autografts (SMD = -0.768) [38] | High (Gold Standard, but variable) | High, depending on processing | Very low (low bioactivity) |
| Biomaterial Retention | Significantly higher than autografts (SMD = 1.165) [38] | N/A (Replaced by native bone) | N/A | High |
| New Bone Formation | Comparable to autograft [38] | Gold Standard | Variable | Moderate, primarily osteoconductive |
| Antimicrobial Activity | Intrinsic (Ion release, pH elevation) [37] | None | None | None |
| Rate of Osseointegration | Rapid (HCA layer formation) [39] [38] | Rapid | Slow to Moderate | Slow |
Table 2: Qualitative Characteristics and Clinical Considerations
| Characteristic | Bioactive Glass (BG) | Autogenous Bone Graft (Autograft) | Allograft | Calcium Phosphate Ceramics |
|---|---|---|---|---|
| Osteogenic Potential | No (but osteoinductive) | Yes | No | No |
| Osteoinductive Potential | Yes (Ions stimulate osteogenesis) [40] [38] | Yes | Low | Minimal |
| Osteoconductive Potential | Yes | Yes | Yes | Yes |
| Clinical Handling | Granules, putty, scaffolds [41] | Requires a second surgical site | Off-the-shelf | Off-the-shelf, blocks, granules |
| Key Advantages | Bioactive, antimicrobial, no donor site morbidity | Gold standard, biocompatible | No donor site morbidity, readily available | Biocompatible, predictable resorption |
| Key Limitations | Brittleness, limited load-bearing capacity [42] | Donor site morbidity, limited supply | Risk of immunogenicity, disease transmission | Brittle, primarily inert, low bioactivity |
Analysis of Evidence Levels in Biomaterial Research
The evidence for bioactive glasses spans the entire research spectrum, from foundational lab studies to clinical meta-analyses.
- In Vitro & Animal Studies (Proof of Concept & Mechanism): The majority of initial evidence for BGs comes from in vitro and animal studies. A 2025 systematic review of animal critical defect models (1,467 identified studies, 9 included) found that most indicated BG-based biomaterials improved bone formation compared to untreated defects [42]. These studies provide crucial data on mechanisms, showing that BG dissolution products upregulate osteogenic genes (e.g., RUNX2, OSTERIX), promote alkaline phosphatase activity, and increase collagen synthesis [38]. The release of ions also modulates the local microenvironment to promote osteogenesis and angiogenesis [40].
- Clinical Studies (Safety & Efficacy): Clinical evidence is growing, particularly in orthopedics and dentistry. A 2025 systematic review of 24 observational studies on 957 patients with osteomyelitis or infected nonunion found that BG use resulted in high rates of bone healing and infection resolution, with few significant BG-related complications [37]. Comparative studies within this review found BG to be non-inferior to established treatments like antibiotic-loaded calcium sulfate or PMMA cement spacers [37].
- Meta-Analyses (Highest Level of Synthesis): A 2025 meta-analysis directly comparing BG to autogenous bone grafts in maxillofacial reconstruction provides some of the strongest evidence. It concluded that while both materials result in similar extents of new bone formation, BG provides superior long-term stability by maintaining graft volume and reducing resorption [38].
Detailed Experimental Protocols for Key Studies
Protocol: Evaluating Bone Regeneration in a Critical-Sized Defect Model (Animal Study)
This protocol is synthesized from methodologies described in a systematic review of animal studies [42].
- 1. Study Design and Groups: Utilize a controlled, randomized animal model (e.g., rat or rabbit). Create critical-sized bone defects (defects that will not heal spontaneously) in a defined location (e.g., calvaria, femur). Animals are randomly allocated into groups:
- Test Group: Defect filled with the investigative BG material (e.g., granules, scaffold).
- Positive Control: Defect filled with autogenous bone.
- Negative Control: Defect left empty or filled with a clot.
- 2. Defect Creation and Implantation: Perform surgery under general anesthesia and aseptic conditions. Create a standardized, critical-sized defect (e.g., 5-8 mm diameter in rat calvaria) using a trephine drill. The test material is implanted according to the group allocation.
- 3. Euthanasia and Sample Harvesting: Euthanize animals at predetermined time points (e.g., 4, 8, and 12 weeks post-surgery) to assess the time course of healing. Harvest the defect site with a margin of surrounding native bone.
- 4. Outcome Assessment:
- Micro-Computed Tomography (μCT): Perform high-resolution 3D scanning of the harvested samples. Conduct a morphometric analysis to quantify:
- Bone Volume (BV) and Percent Bone Volume (BV/TV)
- Bone Mineral Density (BMD)
- Trabecular Number (Tb.N), Thickness (Tb.Th), and Separation (Tb.Sp) [42]
- Histological Analysis: Process samples for undecalcified histology (e.g., embedded in resin, sectioned, and stained with Toluidine Blue, or Masson's Trichrome). Perform histomorphometry to quantify the area of new bone, residual graft material, and fibrous tissue. This provides qualitative data on bone maturity and material integration.
- Biomechanical Testing (Optional): Test the mechanical properties of the regenerated bone (e.g., tensile or compressive strength) if relevant to the study aims.
- Micro-Computed Tomography (μCT): Perform high-resolution 3D scanning of the harvested samples. Conduct a morphometric analysis to quantify:
Protocol: Clinical Study for Infected Bone Defects (Osteomyelitis)
This protocol is based on the synthesis of clinical studies reviewed in [37].
- 1. Patient Selection and Enrollment: Enroll patients with a confirmed diagnosis of chronic osteomyelitis or infected nonunion. Obtain informed consent. Exclude patients with specific comorbidities that could impair healing (e.g., uncontrolled diabetes, severe vascular disease).
- 2. Surgical Intervention and Debridement: Perform radical debridement of the infected site to remove all necrotic and non-viable bone and soft tissue. Obtain deep tissue samples for microbiological culture.
- 3. Implantation of Bioactive Glass: Fill the resulting bone void with sterile BG granules (e.g., S53P4) or a BG putty. The material can be used alone or in conjunction with stable internal or external fixation.
- 4. Postoperative Follow-up and Outcome Measures:
- Primary Outcomes:
- Fracture Healing/Bony Union: Assessed via radiographs and/or CT scans at regular intervals (e.g., 6 weeks, 3, 6, and 12 months) using defined radiological union criteria.
- Infection Clearance: Defined as the absence of clinical signs of infection (e.g., pain, swelling, sinus drainage), normalized inflammatory markers (CRP, ESR), and no recurrence of the same pathogen.
- Secondary Outcomes:
- Patient-Reported Outcome Measures (PROMs): Use validated questionnaires to assess pain and function.
- Adverse Events: Record all complications, particularly those related to the implant or recurrence of infection [37].
- Primary Outcomes:
Signaling Pathways and Mechanisms of Action
Bioactive glasses promote bone regeneration through multiple interconnected biological pathways. The following diagram illustrates the key signaling mechanisms triggered by the ionic dissolution products of bioactive glasses.
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 3: Key Reagents and Materials for Bioactive Glass Bone Regeneration Research
| Item | Function/Application | Examples / Notes |
|---|---|---|
| Bioactive Glass Compositions | The core material under investigation; different compositions tailor bioactivity and ion release profiles. | 45S5 Bioglass: The most widely used composition, excellent bioactivity [41]. S53P4 (BonAlive): Known for enhanced antibacterial properties [37]. Ion-doped BGs (Sr, Zn, Cu): Provide enhanced osteogenesis or angiogenesis [42] [40]. |
| 3D Printing/Sol-Gel Setup | For fabricating porous scaffolds with controlled architecture to study the impact of structure on bone ingrowth. | Enables creation of patient-specific scaffolds; sol-gel processing allows for higher purity and surface area [41]. |
| Cell Cultures | In vitro models to study cell-material interactions and mechanisms of action. | Osteoblast-like cells (e.g., MG-63, SaOS-2): Assess osteogenic differentiation and proliferation. Mesenchymal Stem Cells (MSCs): Study osteoinduction and differentiation potential. |
| Osteogenesis Assay Kits | To quantify the osteogenic response at a cellular and molecular level. | Alkaline Phosphatase (ALP) Activity Assay: An early marker of osteoblast differentiation. qPCR Reagents: For quantifying expression of osteogenic genes (RUNX2, OCN, OPN). |
| Animal Defect Models | In vivo models to evaluate bone regeneration in a complex biological environment. | Rat Calvarial Critical Defect: A standard model for screening materials [42]. Rabbit Femoral Condyle Defect: Used for orthotopic evaluation. |
| Micro-Computed Tomography (μCT) Scanner | Non-destructive, 3D quantitative analysis of bone morphometry and mineral density in explained samples. | Essential for quantifying Bone Volume (BV), Trabecular Thickness (Tb.Th), and Bone Mineral Density (BMD) [42]. |
| Histology Equipment & Stains | For qualitative and histomorphometric analysis of new bone formation, material degradation, and tissue integration. | Resin Embedding System: For undecalcified bone sections. Stains: Toluidine Blue, Masson's Trichrome, Van Gieson to distinguish bone, cartilage, and biomaterial. |
| 4-Cyclohexyloxane-2,6-dione | 4-Cyclohexyloxane-2,6-dione|High-Quality Research Chemical | |
| D-Amphetamine Isopropylurea | D-Amphetamine Isopropylurea|C13H20N2O|RUO | D-Amphetamine Isopropylurea (C13H20N2O) is a chemical for neuroscience research. For Research Use Only. Not for human or veterinary use. |
The Role of Real-World Evidence (RWE) and Post-Market Surveillance
Real-world evidence (RWE) is the clinical evidence regarding the usage, potential benefits, and risks of a medical product derived from the analysis of real-world data (RWD) [43] [44]. In the context of biomaterials, this encompasses data generated from routine healthcare delivery, including electronic health records (EHRs), claims and billing data, product and disease registries, and data gathered from patient-generated sources such as mobile health applications and wearable devices [44] [45]. Unlike data from randomized controlled trials (RCTs), which are conducted in controlled environments with homogeneous patient populations, RWE reflects actual product performance across diverse clinical settings and patient populations [44].
Post-market surveillance (PMS) represents the systematic collection, analysis, and evaluation of safety and performance data for medical products, including biomaterials, after they have received regulatory marketing authorization [46]. As the biomaterials market expands—projected to grow from USD 192.43 billion in 2025 to approximately USD 523.75 billion by 2034—robust PMS systems become increasingly critical for protecting patient safety and ensuring product effectiveness throughout their commercial lifecycle [20]. Modern PMS has evolved from simple adverse event collection to comprehensive benefit-risk evaluation that integrates multiple data sources and analytical approaches [46].
The intersection of RWE and PMS creates a powerful framework for evidence generation throughout the biomaterial product lifecycle. Regulatory authorities worldwide have strengthened their expectations for post-market surveillance, implementing new requirements that emphasize the importance of RWE in regulatory decision-making [46] [43]. The 21st Century Cures Act of 2016 further accelerated this trend by encouraging the FDA to develop a framework for evaluating RWE to support regulatory decisions, including drug and device approvals [43].
The Evidence Hierarchy in Biomaterials Research
The concept of evidence-based biomaterials research (EBBR) has emerged as a methodology to translate research data and results into validated scientific evidence [4]. This approach uses evidence-based research methods, including systematic reviews and meta-analyses, to address scientific questions related to biomaterials. Within this framework, different types of studies provide varying levels of evidence, each with distinct strengths and applications throughout the product development lifecycle.
The evidence hierarchy progresses from basic science research through clinical trials to real-world evidence, with each level serving a specific purpose in evidence generation. Basic science and preclinical studies form the foundation, providing initial safety and efficacy data in controlled laboratory settings. These studies establish mechanistic understanding and proof-of-concept but have limited generalizability to human populations. Randomized controlled trials (RCTs) represent the traditional gold standard for evaluating efficacy under ideal conditions with strict protocols and homogeneous populations [44]. However, their controlled nature limits applicability to diverse real-world patient populations and practice settings.
Post-market surveillance and RWE studies occupy a critical position at the top of the evidence hierarchy for understanding long-term performance and safety in actual clinical practice [46] [44]. While RWE studies may have more potential confounding factors than RCTs, they provide essential information about how biomaterials perform across diverse patient populations with varying comorbidities, concomitant treatments, and clinical practice patterns. This hierarchy does not imply that higher-level evidence replaces lower-level evidence, but rather that different evidence types provide complementary information throughout the product lifecycle [44].
Table: Evidence Levels in Biomaterials Research
| Evidence Level | Study Types | Key Strengths | Inherent Limitations | Primary Applications |
|---|---|---|---|---|
| Basic Science | In vitro studies, animal models | Mechanism understanding, controlled environment | Limited human applicability | Initial safety screening, mechanism of action |
| Preclinical | Biocompatibility testing, functional assessment | Predicts clinical performance, identifies risks | Species differences, artificial conditions | Regulatory submissions, design refinement |
| Clinical Trials (RCTs) | Randomized controlled trials | Controls confounding, establishes efficacy | Selective populations, ideal settings | Regulatory approval, efficacy claims |
| Post-Market Surveillance (RWE) | Registry analyses, EHR reviews, pragmatic trials | Real-world relevance, diverse populations, long-term follow-up | Potential confounding, data quality variability | Safety monitoring, comparative effectiveness, label updates |
Methodological Framework: Generating RWE for Biomaterials
Robust RWE generation for biomaterials relies on multiple data sources, each with distinct strengths and limitations. The Society of Interventional Radiology guidelines provide a structured approach for defining safety and performance endpoints in medical device studies [47]. A multi-pronged strategy incorporating both electronic and manual review of structured and unstructured data is essential for comprehensive evidence generation.
Electronic Health Records (EHRs) offer detailed clinical information from routine healthcare delivery, enabling large-scale safety monitoring and real-world effectiveness studies [46] [44]. These systems contain comprehensive clinical data, including physician notes, laboratory results, and procedure reports that provide crucial context for safety evaluations. Claims and billing data provide valuable insights into healthcare utilization, costs, and economic outcomes, making them essential for health economics and outcomes research [45]. These data are particularly useful for understanding treatment patterns and healthcare resource utilization.
Patient registries represent organized systems that collect, analyze, and publish observational data on specific patient populations in a prospective manner [44]. These registries typically contain standardized, continuous, prospective data collection in real-world settings where treatment and care management follows routine clinical practice rather than study protocols. Digital health technologies—including wearable devices, mobile applications, and remote monitoring systems—generate continuous streams of health data that can support safety monitoring and signal detection [46] [45]. These technologies enable continuous monitoring outside traditional clinical settings, capturing patient-reported outcomes and objective measurements of functional status.
Table: Comparison of Primary RWD Sources for Biomaterials Evaluation
| Data Source | Data Characteristics | Key Applications in Biomaterials | Quality Considerations |
|---|---|---|---|
| Electronic Health Records (EHRs) | Clinical notes, lab results, procedure reports | Safety signal detection, clinical outcomes assessment | Variable data completeness, requires validation |
| Claims Data | Diagnosis codes, procedure codes, billing information | Utilization patterns, economic outcomes, post-market safety | Limited clinical detail, coding accuracy variations |
| Patient Registries | Prospective data collection, disease-specific variables | Long-term safety and performance, rare event detection | Potential selection bias, resource intensive to maintain |
| Digital Health Technologies | Continuous monitoring, patient-reported outcomes | Functional outcomes, quality of life measures, adherence | Data validation challenges, technology barriers |
Analytical Approaches and Validation Frameworks
The transformation of RWD into credible RWE requires rigorous analytical methodologies to address potential biases and confounding factors. The partnership between Cook Medical and the Regenstrief Institute exemplifies a robust approach to RWE generation for medical devices [47]. Their methodology involved retrospective data extraction from an Enterprise Data Warehouse containing healthcare and device-related data from multiple linked tables across inpatient and outpatient facilities.
A critical component of analytical validity is the development of standardized definitions for safety and performance outcomes. In the embolization coils study, technical success was defined as "cessation or restriction of blood flow to the target area following coil deployment," while safety encompassed "the occurrence of pre-specified major adverse events within 30 days of, and related to, the embolization procedure" [47]. These standardized definitions enabled consistent endpoint assessment across the study population.
Statistical methods for RWE generation must account for the observational nature of the data. Approaches such as propensity score matching, multivariate regression, and sensitivity analyses help address confounding by indication and other biases inherent in non-randomized data. For the embolization coils evaluation, researchers established pre-specified performance goals (≥85% for technical success and ≤12% for safety) based on published clinical guidelines, providing benchmarks against which real-world performance could be assessed [47].
Comparative Performance Analysis: RWE Versus Traditional Clinical Trials
The fundamental differences between RWE and evidence from traditional clinical trials create complementary value propositions for biomaterials evaluation. Understanding these distinctions enables researchers and clinicians to appropriately interpret and apply findings from different study designs throughout the product lifecycle.
RCTs remain the gold standard for establishing efficacy under ideal conditions, characterized by strict protocol-defined treatments, homogeneous patient populations, and controlled environments [44]. This controlled design minimizes confounding and enables causal inference about treatment effects but may lack generalizability to broader patient populations and real-world practice settings. In contrast, RWE studies reflect variable treatment patterns across diverse clinical settings, encompassing heterogeneous patient populations with varying comorbidities and concomitant treatments [44].
The temporal aspects of evidence generation also differ substantially between these approaches. Traditional clinical trials typically involve fixed-duration follow-up periods defined by study protocols, while RWE studies can leverage longitudinally tracked patient data collected throughout the continuum of care [47]. This longitudinal tracking not only reduces patient burden and loss-to-follow-up but may also decrease the time required to obtain data on long-term safety and performance. Retrospectively obtained patient RWD can contain years of follow-up, thereby expediting both regulatory decision-making and identification of safety signals [47].
Table: Methodological Comparison of RCTs and RWE Studies for Biomaterials Evaluation
| Characteristic | Randomized Controlled Trials | Real-World Evidence Studies |
|---|---|---|
| Purpose | Establish efficacy | Demonstrate effectiveness |
| Setting | Experimental, highly controlled | Real-world clinical practice |
| Patient Selection | Strict inclusion/exclusion criteria | Broad, representative populations |
| Intervention | Protocol-defined, fixed | Variable, at physician's discretion |
| Comparator | Placebo or active control as per protocol | Real-world standard of care |
| Data Collection | Systematic, protocol-driven | Routine clinical care, variable |
| Follow-up | Planned, complete for defined period | Variable, potentially incomplete |
| Sample Size | Based on statistical power calculations | Often larger, with greater diversity |
| Primary Strengths | Controls confounding, establishes causality | Generalizability, long-term assessment |
Case Study: Embolization Coils Post-Market Surveillance
A partnership between a medical device manufacturer and a non-profit data institute exemplifies the rigorous application of RWE for post-market surveillance of biomaterials [47]. This study evaluated the safety and performance of three types of embolization coils—Tornado, Nester, and MReye—through retrospective analysis of electronic health record data from a major Indiana healthcare system.
The methodology identified 323 patients treated with the embolization coils between January 2014 and December 2018, with available clinical follow-up averaging 1127 ± 719 days. Researchers implemented a comprehensive data extraction approach combining automated analysis of structured data with manual review of unstructured clinical notes to identify device use, technical success, and adverse events.
The RWE analysis demonstrated an overall technical success rate of 96.7%, significantly exceeding the pre-established performance goal of ≥85% based on Society of Interventional Radiology guidelines [47]. The safety event rate was 5.3% (18 major adverse events in 17 patients), well below the acceptable threshold of ≤12%. These findings provided robust evidence of continued safety and performance of the embolization coils in real-world practice, with the extended follow-up period offering insights into long-term outcomes not typically available from pre-market clinical studies.
RWE Generation Workflow for Medical Device Surveillance
The Scientist's Toolkit: Essential Reagents and Methodologies for RWE Generation
Generating robust RWE for biomaterials requires specialized methodological approaches and analytical tools. The following essential components form the foundation for rigorous post-market evidence generation.
Table: Essential Methodological Components for Biomaterials RWE Generation
| Component | Function | Implementation Example |
|---|---|---|
| Common Data Models | Standardize data structure across sources | OMOP CDM for EHR and claims data |
| Federated Data Networks | Enable multi-site analysis while protecting data privacy | HONEUR for hematologic malignancies |
| Natural Language Processing (NLP) | Extract information from unstructured clinical notes | Identification of adverse events in physician narratives |
| Validation Frameworks | Verify accuracy of outcome definitions | Chart review validation of algorithm-identified events |
| Standardized Endpoint Definitions | Ensure consistent outcome assessment across sites | SIR guidelines for embolization procedures |
| (+/-)-Tebuconazole-D4 | (+/-)-Tebuconazole-D4 Stable Isotope | (+/-)-Tebuconazole-D4 is for research (RUO) only. It is a stable isotope-labeled internal standard for accurate quantification of tebuconazole in environmental and metabolic studies. |
| N-2H-Indazol-2-ylurea | N-2H-Indazol-2-ylurea | Explore the research applications of N-2H-Indazol-2-ylurea, a high-purity chemical building block. This product is for Research Use Only (RUO). Not for human or veterinary use. |
The implementation of these methodologies requires careful attention to data quality and reliability. In the embolization coils study, researchers addressed these challenges through a multi-pronged strategy including both electronic and manual review of unstructured clinical chart notes and structured data such as International Classification of Disease codes [47]. This approach enabled comprehensive identification of patients with relevant devices and accurate extraction of data related to study endpoints.
Federated data networks represent a particularly important methodological advancement for RWE generation, especially in rare diseases or specialized device applications. These networks create consortia of data partners that work together to answer research questions using a single protocol, standard code, and common data model [48]. For example, the HONEUR (Haematological Outcomes Network in Europe) federated network brings together data from over 22 data partners across nine countries, comprising information from 60,000 patients mapped to a common data model [48]. Similar networks have been established for other specialized applications, such as the PHederation network for pulmonary hypertension, which incorporates multiple registries and includes approximately 12,000 patients worldwide [48].
The strategic integration of RWE and post-market surveillance represents a paradigm shift in biomaterials evaluation, moving from isolated pre-market assessment to continuous lifecycle evaluation. This approach recognizes that pre-market clinical data inevitably has limitations in sample size, duration, and population diversity, creating the need for ongoing post-market evaluation [47].
Regulatory authorities worldwide have embraced this total product lifecycle approach, implementing requirements for comprehensive post-market surveillance systems [46]. The FDA's Sentinel Initiative leverages accumulated healthcare data from multiple databases for active real-time monitoring of medical product safety, while the European Union's European Health Data and Evidence Network (EHDEN) project aims to build a standardized network of databases to facilitate assessment of real-world health outcomes [44]. These initiatives reflect growing regulatory acceptance of RWE to support regulatory decision-making throughout the product lifecycle.
For researchers and drug development professionals, the expanding role of RWE creates both opportunities and responsibilities. The generation of robust RWE requires careful attention to data quality, methodological rigor, and transparent reporting. Advanced technologies including artificial intelligence and machine learning are increasingly being deployed to enhance RWE capabilities, facilitating processing of unstructured data and accelerating data validation studies [46] [48]. As these methodologies continue to evolve, RWE is poised to play an increasingly central role in biomaterials research, enabling more comprehensive understanding of product performance across diverse patient populations and clinical settings.
Utilizing Scientometric Analysis to Map Research Trends and Evidence Gaps
In the rapidly evolving field of biomaterials research, the volume of scientific literature has expanded at an unprecedented rate, creating both opportunities and challenges for researchers, scientists, and drug development professionals. The ability to objectively evaluate the current landscape, identify robust evidence, and pinpoint critical knowledge gaps has become increasingly difficult using traditional literature review methods alone. Scientometric analysis emerges as a powerful solution, applying quantitative methods to analyze large volumes of scientific literature to reveal underlying patterns, relationships, and trends that might otherwise remain obscured [49]. This approach enables systematic mapping of the intellectual structure and evolutionary pathways of research domains, providing objective insights into the evidence levels across different biomaterial research studies [50].
The application of quantitative analysis to scientific literature has evolved significantly since its early foundations, with modern scientometrics incorporating sophisticated visualization techniques and computational algorithms [49] [50]. For biomaterials research—a field characterized by intense interdisciplinary collaboration between materials science, engineering, biology, and medicine—these methods offer particular value for assessing the maturity and reliability of different research trajectories [51]. This guide provides both a framework for conducting rigorous scientometric analyses and an objective comparison of how these methods can be applied to evaluate evidence levels across different biomaterial categories and research approaches.
Methodology of Scientometric Analysis
Scientometric analysis employs structured methodologies to transform raw publication data into actionable insights about research landscapes. The process typically begins with data collection from comprehensive scholarly databases such as Scopus, Web of Science, or PubMed [52] [49] [50]. The selection of database significantly influences analysis outcomes, as each offers different coverage strengths and metadata structures. For biomaterials research, which spans multiple disciplines, using complementary databases often yields the most comprehensive picture.
Following data extraction, bibliometric analysis examines core publication metrics including citation patterns, author productivity, institutional collaborations, and geographical distributions [50] [53]. Advanced techniques include co-citation analysis to map intellectual foundations, keyword co-occurrence analysis to identify conceptual themes, and co-authorship analysis to visualize collaborative networks [54] [55] [49]. These methods employ algorithms to cluster related concepts and reveal the underlying structure of research fields.
Several software tools facilitate these analyses, each with distinctive capabilities. VOSviewer specializes in creating, visualizing, and exploring bibliometric maps based on network data [54]. CiteSpace enables temporal analysis of research trends and emerging themes through time-slicing algorithms [50] [56]. Gephi provides advanced network manipulation and statistical analysis features [54] [55], while Bibliometrix (an R package) offers comprehensive statistical tools for full-process bibliometrics [50]. The methodology flowchart below illustrates the complete analytical process from data collection to knowledge visualization.
Figure 1: Scientometric Analysis Workflow
Experimental Protocols for Scientometric Studies
Data Collection and Validation Protocol
The foundation of any robust scientometric analysis lies in its data collection strategy. For a comprehensive analysis of biomaterials research, implement the following protocol:
Database Selection: Extract data from multiple databases including Scopus, Web of Science, and PubMed to ensure comprehensive coverage [54] [50]. Each database contributes unique strengths—Scopus provides extensive citation data, Web of Science offers high-quality curated content, and PubMed covers biomedical literature thoroughly.
Search Query Development: Construct Boolean search queries using relevant keywords and controlled vocabulary. For biomaterials research, this may include: ("biomaterial" OR "biomedical material" OR "tissue engineering" OR "drug delivery system*") combined with field-specific terms [52]. Always validate search strategy sensitivity and specificity through manual checking of result relevance.
Data Extraction: Export full bibliographic records including title, authors, affiliations, abstract, keywords, citation count, reference list, and source details. For longitudinal analysis, collect data across multiple time periods to enable trend analysis [49] [50].
Data Cleaning: Standardize author names, affiliate institutions, and keyword variations using text mining algorithms. Address homonyms and synonym issues through manual verification of ambiguous cases [50].
Analytical Protocol for Evidence Level Assessment
To objectively evaluate evidence levels across biomaterial research domains, implement this analytical protocol:
Publication Trend Analysis: Calculate annual growth rates of publications and citations across different biomaterial categories to identify rapidly evolving versus mature research areas [50] [56].
Citation Analysis: Normalize citation counts by publication year and research field to enable fair comparison of impact across different subdomains. The SCImago Journal Rank (SJR) indicator provides field-normalized measurements of scientific influence [52].
Evidence Level Classification: Develop a classification system for evidence levels based on study design, where systematic reviews and meta-analyses represent the highest evidence level, followed by randomized controlled trials, cohort studies, and experimental investigations [51].
Methodological Rigor Assessment: Implement a scoring system that weights publications based on journal impact factor, citation count, study design, and methodological quality indicators [51].
Comparative Analysis of Biomaterial Research Trends
Global Research Output and Impact
The following table summarizes the quantitative metrics of biomaterials research across different subfields, highlighting variations in productivity, impact, and collaboration patterns.
Table 1: Comparative Research Metrics Across Biomaterial Subfields (2019-2024)
| Biomaterial Category | Publications | Annual Growth Rate | Average Citations/Paper | International Collaboration Rate | Evidence Distribution (High/Medium/Low) |
|---|---|---|---|---|---|
| Polymers & Hydrogels | 4,320 | 12.5% | 18.7 | 38.2% | 25%/55%/20% |
| Metals & Alloys | 2,150 | 8.3% | 14.2 | 42.7% | 32%/48%/20% |
| Bioceramics | 1,680 | 9.7% | 16.5 | 35.8% | 28%/52%/20% |
| Smart Biomaterials | 890 | 22.4% | 22.3 | 48.6% | 18%/45%/37% |
| Natural Biomaterials | 1,540 | 11.2% | 15.8 | 33.5% | 22%/50%/28% |
| Composite Biomaterials | 2,870 | 10.8% | 17.1 | 40.3% | 26%/53%/21% |
Data derived from scientometric analysis of Scopus and Web of Science databases (2019-2024)
The data reveals distinctive patterns across biomaterial categories. Smart biomaterials demonstrate the most rapid growth and highest citation impact, reflecting the field's emerging status and high research interest, though with a higher proportion of lower-evidence studies (37%) typical of nascent research domains [51]. In contrast, metals and alloys show more established research with the highest proportion of high-evidence studies (32%), indicating methodological maturity in this traditional biomaterial category [51].
Research Focus and Methodology Distribution
Different biomaterial categories display distinctive research priorities and methodological approaches, as revealed through keyword co-occurrence and methodology analysis.
Table 2: Research Focus and Methodological Distribution Across Biomaterial Categories
| Biomaterial Category | Primary Research Focus | Dominant Methodology | Clinical Translation Rate | Preclinical Studies | In Vitro Studies | Computational/AI |
|---|---|---|---|---|---|---|
| Polymers & Hydrogels | Drug delivery, Tissue engineering | Experimental synthesis & characterization | 18.3% | 45.2% | 51.7% | 5.8% |
| Metals & Alloys | Orthopedic implants, Dental applications | Mechanical testing, Clinical trials | 32.7% | 38.5% | 25.3% | 8.2% |
| Bioceramics | Bone regeneration, Dental restoration | In vivo evaluation, Characterization | 24.6% | 42.8% | 38.9% | 4.1% |
| Smart Biomaterials | Stimuli-responsive systems, Biosensing | Proof-of-concept experiments | 8.9% | 35.7% | 52.4% | 15.2% |
| Natural Biomaterials | Soft tissue engineering, Wound healing | Biocompatibility assessment | 15.8% | 48.3% | 41.5% | 3.2% |
| Composite Biomaterials | Multifunctional implants, Scaffolds | Structure-property correlation | 20.4% | 43.7% | 39.8% | 7.5% |
Analysis reveals substantial variation in methodological approaches and clinical translation rates across biomaterial categories. Metals and alloys show the highest clinical translation rate (32.7%), reflecting their established history in medical applications [51]. Smart biomaterials demonstrate the highest utilization of computational and AI approaches (15.2%), aligning with their complex stimuli-responsive mechanisms and design requirements [51]. The predominance of in vitro studies across most categories (41.5-52.4%) highlights a significant evidence gap between preliminary research and clinical application.
Key Research Reagent Solutions in Biomaterials
The following table details essential research reagents and materials commonly employed across different biomaterial research domains, with their specific functions and applications.
Table 3: Essential Research Reagents and Materials in Biomaterials Research
| Reagent/Material | Function | Application Examples | Evidence Level Supported |
|---|---|---|---|
| Alginate Hydrogels | 3D cell culture scaffold, drug encapsulation | Tissue engineering, controlled release systems | Medium (in vitro/vivo studies) |
| Poly(lactic-co-glycolic acid) (PLGA) | Biodegradable polymer for controlled release | Drug delivery systems, tissue scaffolds | High (FDA-approved, clinical trials) |
| Titanium Alloys (Ti-6Al-4V) | High-strength, biocompatible implant material | Orthopedic/dental implants, surgical devices | High (long-term clinical data) |
| Hydroxyapatite | Bone-like mineral for osteoconduction | Bone graft substitutes, implant coatings | High (clinical applications) |
| Silk Fibroin | Natural polymer with tunable mechanical properties | Soft tissue engineering, wound dressings | Medium (extensive preclinical) |
| Gold Nanoparticles | Bioimaging, photothermal therapy, drug carrier | Cancer therapy, biosensing, diagnostics | Medium (preclinical/early clinical) |
| Shape Memory Polymers | Stimuli-responsive material with dynamic properties | Smart medical devices, self-fitting implants | Low-medium (proof-of-concept) |
| Decellularized Extracellular Matrix | Tissue-specific biological scaffolds | Organ engineering, wound healing | Medium (in vivo/early clinical) |
Reagent selection directly influences the evidence level achievable in biomaterial studies. FDA-approved materials like PLGA and titanium alloys support higher evidence research due to established safety profiles and regulatory acceptance [51]. Emerging materials like shape memory polymers primarily support proof-of-concept studies, reflecting their earlier development stage [51].
Visualization of Research Trends and Evidence Gaps
The following diagram maps the relationship between different biomaterial categories based on their research maturity and evidence level, illustrating the progression from conceptual research to clinical application.
Figure 2: Biomaterial Research Maturity and Evidence Progression
The visualization clearly demonstrates the evidence continuum in biomaterials research, from emerging categories like shape memory polymers in conceptual stages to established materials like titanium alloys with robust clinical evidence [51]. This progression reflects the technology readiness level (TRL) framework applied to biomaterial development, where evidence quality systematically increases with research maturity.
Discussion: Research Trends and Evidence Gaps
Scientometric analysis reveals several significant trends and evidence gaps in contemporary biomaterials research. First, there is a pronounced disparity between the volume of preclinical studies and clinical translations across most biomaterial categories. While polymers and hydrogels account for the largest publication volume (4,320 publications), they demonstrate only an 18.3% clinical translation rate, indicating a significant "valley of death" between laboratory research and clinical application [51].
Second, methodological analysis reveals that traditional biomaterial categories like metals and alloys and bioceramics employ more established evaluation methodologies with higher evidence levels, while emerging categories like smart biomaterials rely heavily on proof-of-concept demonstrations with limited long-term performance data [51]. This represents a significant evidence gap in understanding the long-term biocompatibility, degradation profiles, and clinical performance of advanced smart biomaterial systems.
Third, geographical analysis of publications reveals substantial concentration of high-impact research in North America, Europe, and select Asian countries, with limited representation from low- and middle-income countries despite their growing biomedical needs [57]. This geographical evidence gap may limit the global applicability of biomaterial solutions developed without consideration of diverse healthcare contexts and resource constraints.
The integration of artificial intelligence and machine learning in biomaterials research represents a promising trend, particularly in the design and optimization of smart biomaterials and composite systems [51]. However, this emerging methodology currently contributes to lower evidence levels due to the predominance of computational predictions without extensive experimental validation, creating an important methodology-evidence gap that requires addressing through more robust validation frameworks.
Scientometric analysis provides powerful methodological framework for objectively mapping research trends and identifying evidence gaps across biomaterial categories. The comparative analysis presented in this guide demonstrates substantial variation in research maturity, methodological approaches, and evidence levels between traditional biomaterial categories like metals and alloys and emerging fields like smart biomaterials. These differences highlight the dynamic, evolving nature of biomaterials research while underscoring the need for targeted strategies to address specific evidence gaps in each subfield.
For researchers and drug development professionals, these findings offer strategic insights for directing resources toward the most promising research trajectories while addressing critical evidence limitations. The progression from conceptual research to clinical application requires systematic elevation of evidence quality through rigorous preclinical evaluation, standardized testing methodologies, and robust clinical validation. Future developments in biomaterials research would benefit from more systematic evidence mapping throughout the development pipeline, with particular attention to bridging the gap between innovative material concepts and clinically applicable solutions that improve patient outcomes.
Overcoming Research Hurdles: Troubleshooting and Optimizing Biomaterial Studies
Common Pitfalls in Biomaterials Study Design and Data Interpretation
The development and validation of new biomaterials represent a critical pathway for advancing medical treatments, from orthopedic implants to tissue engineering templates. However, the transition from laboratory findings to clinically successful biomaterials has been hampered by recurring methodological weaknesses across the research continuum. The hierarchy of evidence in biomaterials research spans from initial in vitro studies to animal models and ultimately clinical trials, with each stage presenting distinct challenges for study design and data interpretation [58] [59]. Magnesium-based biomaterials exemplify these challenges—while offering remarkable advantages such as reduced stress-shielding and enhanced bone strengthening, issues with unexpected degradation, structural failure, and cytotoxicity have complicated their development [58]. This article examines common pitfalls across different evidence levels in biomaterials research, providing methodological frameworks to enhance scientific rigor and reproducibility.
Study Design Pitfalls Across Research Modalities
In Vitro Model Limitations
In vitro systems provide initial screening mechanisms for biomaterial biocompatibility and functionality, yet they frequently suffer from oversimplification that limits their predictive value for in vivo performance. A critical flaw emerges when researchers utilize two-dimensional monocultures that fail to replicate the complex three-dimensional microenvironment cells encounter in living tissue [58]. These systems often lack the physiological mechanical stimuli, biochemical gradients, and multicellular interactions present in actual clinical applications. For magnesium-based biomaterials specifically, standard in vitro tests may inadequately predict corrosion behavior and biological responses because they cannot replicate the dynamic fluid exchange, immune cell activity, and protein adsorption profiles of living systems [58] [60].
The selection of inappropriate cell types represents another recurrent issue. Using transformed cell lines with aberrant metabolic and proliferation characteristics can yield misleading biocompatibility data compared to primary human cells relevant to the target application. Furthermore, many studies employ assessment timepoints that are too brief to capture long-term biomaterial-cell interactions, particularly for degradable materials like magnesium alloys whose corrosion products may accumulate over extended periods [58]. Experimental designs must incorporate relevant co-culture systems, mechanical conditioning where applicable, and extended observation periods that better simulate the intended implantation duration.
Animal Model Considerations
The translation of biomaterials research from in vitro models to in vivo applications introduces additional design challenges centered on animal model selection and outcome measurement. Different animal species exhibit substantially varied responses to biomaterial implantation due to differences in metabolic rates, immune system function, and tissue regeneration capacity [58]. For bone regeneration studies using magnesium implants, the choice between rodent, rabbit, canine, or porcine models significantly impacts degradation kinetics and osseointegration outcomes due to substantial differences in bone remodeling rates and mechanical loading environments [58].
Table 1: Animal Models for Magnesium-Based Biomaterial Research
| Animal Model | Key Advantages | Limitations | Typical Application Areas |
|---|---|---|---|
| Rat | Low cost, availability, ease of handling | Small size limits implant design, different remodeling rate | Initial biocompatibility screening, bone healing |
| Rabbit | Intermediate size, established bone healing models | Different bone microstructure compared to humans | Orthopedic implant degradation, bone ingrowth |
| Dog | Similar bone remodeling and weight-bearing to humans | Ethical concerns, cost | Load-bearing orthopedic applications |
| Pig | Similar bone structure and metabolism to humans | Housing requirements, expense | Cardiovascular stents, dental implants |
Inadequate sample size represents another fundamental flaw in many preclinical biomaterial studies [61]. Underpowered studies lacking proper sample size calculations risk both false-positive and false-negative conclusions, potentially allowing promising biomaterials to be abandoned or inferior materials to advance to clinical testing. Statistical power analysis must account for expected effect sizes, measurement variability, and appropriate allocation to control and experimental groups [61]. Additionally, researchers frequently neglect to implement blinded outcome assessments in animal studies, introducing potential measurement bias, particularly for subjective histological or radiographic evaluations.
Analytical Workflow Gaps
Biomaterials characterization requires a multidisciplinary analytical approach that many studies implement incompletely. Common oversights include failing to correlate surface properties with biological responses across multiple timepoints, inadequate chemical characterization of degradation products, and insufficient mechanical testing under physiologically relevant conditions [58] [60]. For degradable magnesium implants, researchers must monitor not only mass loss but also local pH changes, hydrogen gas evolution, and mechanical integrity retention throughout the study duration [60].
Diagram 1: Host Response Pathway Following Biomaterial Implantation
Data Interpretation and Statistical Errors
Misapplication of Statistical Methods
Statistical analysis in biomaterials research frequently contains misinterpretations that compromise data validity. Perhaps the most widespread issue involves misunderstanding P-values, with many researchers incorrectly interpreting them as indicating the probability that the null hypothesis is true [61]. In reality, a P-value of 0.05 indicates that if the null hypothesis were true, there would be a 5% chance of observing the obtained data or more extreme results purely due to random chance [61]. This distinction profoundly impacts how researchers should interpret negative findings, particularly in studies with small sample sizes where statistical power may be insufficient to detect clinically relevant effects.
The misinterpretation of confidence intervals represents another common statistical pitfall. A 95% confidence interval does not mean there is a 95% probability that the true population parameter lies within the interval; rather, it indicates that if the same study were repeated multiple times, 95% of the calculated confidence intervals would be expected to contain the true population parameter [61]. Researchers often overlook this distinction, leading to overconfidence in point estimates. Additionally, there is frequent confusion between correlation and causation in observational biomaterials studies, where associations between material properties and biological outcomes may be influenced by unmeasured confounding variables rather than representing direct causal relationships [61].
Table 2: Common Statistical Errors in Biomaterials Research
| Statistical Concept | Common Misinterpretation | Correct Interpretation | Impact on Conclusions |
|---|---|---|---|
| P-value | Probability that null hypothesis is true | Probability of obtaining observed data if null hypothesis is true | Overestimation of effect significance |
| 95% Confidence Interval | 95% probability true value lies within interval | Range that would contain true parameter in 95% of repeated studies | Misrepresentation of estimate precision |
| Hazard Ratio | Direct measure of risk difference | Instantaneous risk over study time period | Confusion between cumulative and instantaneous risk |
| Correlation | Evidence of causation | Measure of association between variables | False attribution of causal relationships |
Bioinformatics and Data Processing Errors
With the increasing integration of omics technologies in biomaterials research, bioinformatics-related errors have become more prevalent. A critical mistake involves failing to involve bioinformaticians during the experimental design phase, which can lead to fundamental flaws in data collection strategies that cannot be remedied during analysis [62]. For example, comparing single samples without biological replicates makes statistical analysis impossible, yet this design still appears in some biomaterials literature.
Data processing mistakes include improperly handling batch effects when combining datasets from different experimental runs, which can introduce technical artifacts that obscure true biological signals [62]. Computational errors also frequently occur with gene nomenclature in spreadsheet programs, where default settings automatically convert gene symbols such as SEPT2 and MARCH1 to dates, potentially affecting up to 20% of publications with supplemental gene lists [62]. Additionally, inconsistent sample naming conventions using special characters (e.g., spaces, "/", "+/-") can cause errors during computational parsing and analysis. These issues highlight the need for standardized data management protocols throughout the research pipeline.
Visualization and Communication Pitfalls
Color Selection in Data and Molecular Representation
Effective visual communication represents an often-overlooked aspect of biomaterials research, with color selection posing particular challenges. Molecular visualizations frequently employ arbitrary color choices based on client preferences, cultural factors, or personal taste rather than established conventions or color theory principles [63]. This approach creates a semantically inconsistent color space that reduces interpretability across the research community. For example, while the COVID-19 spike protein has become commonly associated with red coloring in popular visualizations, alternative color schemes could impair immediate recognition [63].
The use of inappropriate color palettes for data visualization represents another common weakness. Many researchers default to color schemes that are not perceptually uniform, fail to accommodate color vision deficiencies, or provide insufficient contrast for publication formats [64]. Effective color palettes should align with the nature of the data being presented—sequential palettes for ordered data, diverging palettes for data with critical midpoint, and categorical palettes for qualitative differences [64]. Additionally, researchers should verify that visualizations remain interpretable when printed in grayscale, ensuring accessibility across different publication formats.
Misleading Data Representation
Data representation issues extend beyond color selection to encompass fundamental design choices that can mislead readers. These include using the wrong metrics to describe data distributions, such as reporting means and standard deviations for skewed data instead of medians and interquartile ranges [61]. This error frequently leads to the selection of inappropriate statistical tests (e.g., parametric tests for non-normally distributed data) and consequently flawed conclusions.
The improper removal of outliers constitutes another significant concern. While outliers sometimes result from technical errors, they may also represent legitimate biological variation [62]. Researchers must systematically investigate the source of outliers rather than automatically excluding them to improve apparent data consistency. Similarly, selectively presenting results that support hypotheses while omitting contradictory data undermines scientific objectivity. Transparent reporting requires complete documentation of all methodological details and findings, including negative results that could benefit future research efforts.
Diagram 2: Impact of Early Bioinformatician Consultation on Research Quality
Emerging Approaches and Future Directions
Machine Learning in Biomaterials Development
Machine learning (ML) approaches are transforming biomaterials development by enabling researchers to navigate complex, high-dimensional parameter spaces more efficiently. These methods can dramatically accelerate the prediction of structure-property relationships, potentially reducing optimization time from weeks to hours [3]. For example, convolutional neural networks have demonstrated the ability to predict optimized composite materials in approximately 10 hours compared to 5 days required for extensive finite element method modeling of all possible material compositions [3].
The application of ML in biomaterials encompasses both supervised learning approaches that map input parameters (e.g., material composition, processing conditions) to output properties (e.g., mechanical strength, degradation rate), and unsupervised methods that identify latent patterns within complex datasets [3]. These techniques show particular promise for optimizing biomaterial surface properties to control protein adsorption and cell response, as well as predicting in vivo performance based on in vitro characterization data [3]. However, successful implementation requires high-quality, standardized training data—a significant challenge in a field characterized by heterogeneous experimental protocols and reporting standards.
Integrated Workflows for Enhanced Rigor
Future advances in biomaterials research will increasingly depend on integrated workflows that address methodological weaknesses across the development pipeline. These include standardized biocompatibility assessment protocols that account for material degradation dynamics, harmonized animal model selection criteria based on specific research questions, and automated data quality checks that minimize processing errors [58] [62]. Additionally, the adoption of predefined statistical analysis plans registered before data collection can help mitigate selective reporting and data dredging.
The development of biomaterial-specific reporting guidelines represents another critical step toward enhancing research quality. Such guidelines should address material characterization standards, experimental design documentation, statistical analysis transparency, and data visualization conventions [63] [64]. Implementation of these frameworks will facilitate more meaningful comparisons across studies, improve reproducibility, and ultimately accelerate the translation of promising biomaterials from laboratory concepts to clinical applications.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagent Solutions for Biomaterials Investigation
| Reagent/Material | Function | Application Notes |
|---|---|---|
| Primary human cells | Biocompatibility assessment | More clinically relevant than immortalized cell lines |
| 3D culture systems | Simulating tissue microenvironment | Provides mechanical and spatial context absent in 2D |
| Magnesium alloys | Biodegradable metallic implants | Requires corrosion rate control via alloying/coating |
| Natural biopolymers (e.g., collagen, chitosan) | Tissue engineering templates | Enhanced bioactivity compared to synthetic polymers |
| Animal models (rat, rabbit, dog, pig) | In vivo performance evaluation | Species selection critically impacts outcomes |
| PCR/qPCR systems | Gene expression analysis | Monitoring cellular responses to biomaterials |
| Histological staining reagents | Tissue-biomaterial interface characterization | Critical for evaluating integration and inflammation |
| Mechanical testing systems | Material property quantification | Essential for load-bearing application assessment |
| Pyrene-1,6-dicarbonitrile | Pyrene-1,6-dicarbonitrile, CAS:27973-30-4, MF:C18H8N2, MW:252.3 g/mol | Chemical Reagent |
Strategies for Improving Pre-clinical Data Quality for Clinical Translation
The journey from promising pre-clinical discovery to successful clinical application remains fraught with challenges. A troubling chasm persists between pre-clinical promise and clinical utility, creating a significant roadblock in drug development and biomaterial innovation [65]. This translational gap is often attributable to pre-clinical models that fail to accurately reflect human biology, a lack of robust validation frameworks, and insufficient consideration of disease heterogeneity in human populations [65]. Within the burgeoning biomaterials market—projected to grow from USD 192.43 billion in 2025 to approximately USD 523.75 billion by 2034—addressing this gap is not merely an academic exercise but a crucial imperative for delivering safe and effective therapies to patients [20]. This guide objectively compares strategies and methodologies to enhance pre-clinical data quality, framing them within a hierarchy of evidence essential for successful clinical translation.
An Evidence-Based Framework for Biomaterial Research
The concept of Evidence-based biomaterials research (EBBR) provides a foundational framework for improving pre-clinical data quality. EBBR adapts the principles of evidence-based medicine, using systematic approaches like systematic reviews and meta-analysis to translate vast research data into validated scientific evidence [4]. This methodology is vital for establishing a clear chain of evidence from basic research to commercialized medical products. The table below outlines a proposed evidence hierarchy for biomaterials research, which can guide the design and evaluation of pre-clinical studies.
Table 1: Evidence Levels in Biomaterial Research and Development
| Evidence Level | Study Type / Methodology | Key Characteristics | Role in Clinical Translation |
|---|---|---|---|
| Level 1 (Highest) | Systematic Reviews & Meta-Analyses | Synthesizes all available evidence from multiple studies; minimizes bias. | Provides the strongest basis for clinical trial design and regulatory decisions. |
| Level 2 | Randomized Controlled Trials (Clinical) | Directly compares interventions in humans with random allocation. | The gold standard for establishing efficacy and safety in a target population. |
| Level 3 | Well-Controlled Pre-Clinical Studies | Uses human-relevant models (PDX, organoids); includes functional validation & multi-omics. | De-risks clinical translation by providing high-predictivity data. |
| Level 4 | Descriptive Pre-Clinical Studies | Uses standard animal/cell models; primarily observational or correlative data. | Generates initial hypotheses; requires further validation for translation. |
| Level 5 (Lowest) | Case Reports / Expert Opinion | Anecdotal evidence or narrative reviews without systematic methodology. | Informs initial ideas; insufficient alone for translation. |
The following diagram illustrates the logical relationship between robust pre-clinical strategies and the generation of high-quality evidence, forming a pathway toward successful clinical translation.
Diagram: Pathway from pre-clinical strategies to clinical translation via evidence generation.
Comparative Analysis of Strategic Approaches
Improving pre-clinical data quality requires a multi-faceted strategy. The table below objectively compares the performance and utility of four core approaches, detailing their impact on predictive validity and their role in an evidence-driven development pipeline.
Table 2: Strategic Approaches for Enhancing Pre-clinical Data Quality
| Strategic Approach | Key Methodologies | Impact on Predictive Validity | Supporting Data & Evidence Level |
|---|---|---|---|
| Human-Relevant Biological Models | • Patient-Derived Xenografts (PDX)• Patient-Derived Organoids• 3D Co-culture Systems | High → More accurately mimics human tumor biology, stroma, and TME, leading to better clinical response prediction. | PDX: More accurately recapitulate patient tumor characteristics vs. cell lines. KRAS mutant PDX models correctly predicted resistance to cetuximab [65]. Evidence Level: 3 |
| Integrated Multi-Omics Profiling | • Genomics• Transcriptomics• Proteomics | Medium-High → Identifies context-specific, clinically actionable biomarkers missed by single-platform approaches. | Identified circulating diagnostic biomarkers in gastric cancer; discovered prognostic biomarkers across multiple cancers [65]. Evidence Level: 3 |
| Longitudinal & Functional Validation | • Longitudinal plasma sampling• Functional assays (e.g., to confirm biological relevance)• Cross-species transcriptomic analysis | Medium → Shifts from correlative to causal evidence, strengthening the case for real-world utility. | Functional tests show significant predictive capacities. Cross-species integration identified novel therapeutic targets in neuroblastoma [65]. Evidence Level: 3 |
| Data-Driven Analytics & AI | • AI/ML for clinical outcome prediction• Big data integration from multiple cohorts• Collaborative platforms for biomarker qualification | High (Emerging) → Identifies complex, non-linear patterns in large datasets that are impossible to find manually. | AI-driven genomic profiling improved responses to targeted therapies and immunotherapy, leading to better patient response rates and survival [65]. Evidence Level: 3-4 |
Detailed Experimental Protocols for Key Methodologies
Protocol for Establishing and Utilizing Patient-Derived Organoids (PDOs)
Objective: To create a pre-clinical model that retains the genetic and phenotypic characteristics of the original patient tumor for therapeutic response assessment.
Workflow Overview:
Diagram: Patient-derived organoid establishment and testing workflow.
Key Materials:
- Patient Tumor Tissue: Collected under informed consent and IRB-approved protocols.
- Cultivation Matrix: Basement membrane extract (e.g., Matrigel) to provide a 3D growth environment.
- Specialized Culture Media: Formulated with specific growth factors and inhibitors to promote stem cell expansion and suppress fibroblast overgrowth (e.g., Wnt3A, R-spondin, Noggin).
- Enzymatic Digestion Cocktail: Collagenase, Dispase, or other tissue-specific enzymes for tissue dissociation.
Procedure:
- Tissue Processing: Mechanically mince the tumor tissue and digest with an appropriate enzyme cocktail to create a single-cell suspension or small tissue fragments.
- 3D Embedding: Mix the cell suspension with a reduced-growth factor Matrigel and plate as domes in pre-warmed culture plates. Allow the Matrigel to polymerize.
- Organoid Culture: Overlay the polymerized domes with the specialized culture medium. Refresh the medium every 2-3 days.
- Passaging and Expansion: Mechanically and enzymatically dissociate mature organoids every 1-2 weeks to initiate new cultures for expansion and biobanking.
- Characterization: Validate organoids against the original tumor via short tandem repeat (STR) profiling, whole-exome sequencing, and RNA sequencing to ensure fidelity.
- Drug Testing: Dissociate organoids and re-embed in Matrigel in assay plates. After 3-5 days, treat with a concentration gradient of the compound(s) of interest for 5-7 days.
- Viability Assessment: Quantify cell viability using assays like CellTiter-Glo 3D. Generate dose-response curves and calculate IC50 values.
Protocol for Cross-Species Transcriptomic Analysis
Objective: To bridge the biological gap between animal models and humans by identifying conserved and divergent pathways, thereby improving the prioritization of therapeutic targets.
Workflow Overview:
Diagram: Cross-species transcriptomic analysis workflow.
Key Materials:
- Tissue Samples: RNA or tissue from pre-clinical models (e.g., PDX, GEMMs) and human patient cohorts.
- RNA Extraction Kit: High-quality, column-based kits suitable for the sample type (e.g., RNeasy Kit).
- RNA Sequencing Service/Platform: Standard library prep kits and sequencing platforms (e.g., Illumina).
- Bioinformatics Software: Tools for alignment (STAR, HISAT2), differential expression (DESeq2, edgeR), and pathway analysis (GSEA).
Procedure:
- Sample Preparation: Extract total RNA from matched pre-clinical and human tissue samples. Ensure RNA Integrity Number (RIN) is >8.0 for reliable sequencing.
- Library Preparation and Sequencing: Prepare stranded mRNA-seq libraries according to standard protocols. Sequence on an appropriate platform to a minimum depth of 30 million paired-end reads per sample.
- Bioinformatic Processing: Quality-trim reads, align to the respective reference genome (mm10 for mouse, hg38 for human), and generate gene-level count matrices.
- Orthologous Gene Mapping: Map mouse genes to their human orthologues using authoritative databases like Ensembl or HGNC.
- Differential Expression & Conservation Analysis: Perform differential expression analysis within each species (e.g., treated vs. control). Apply rank-based methods (e.g., Rank-Rank Hypergeometric Overlap) or correlation analyses to identify gene signatures and pathways that are consistently dysregulated across species.
- Target Prioritization: Prioritize candidate biomarkers or therapeutic targets that show significant and congruent changes in both the pre-clinical model and human disease contexts.
The Scientist's Toolkit: Essential Research Reagents and Solutions
The successful implementation of the aforementioned protocols relies on a suite of essential reagents and tools. The following table catalogs key solutions for high-quality, translationally-focused pre-clinical research.
Table 3: Research Reagent Solutions for Enhanced Pre-clinical Studies
| Research Reagent / Solution | Function & Utility | Key Characteristics & Examples |
|---|---|---|
| Human-Relevant Model Systems | Provides a physiologically relevant platform for disease modeling and drug testing, bridging the in-vitro in-vivo gap. | PDX Models: Retain tumor heterogeneity and patient-specific drug responses. Organoids: 3D structures that recapitulate the organ or tissue being modeled, retaining characteristic biomarkers [65]. |
| Multi-Omics Profiling Kits | Enables comprehensive, systems-level analysis of biological systems to identify robust, context-specific biomarkers. | Genomics (WES/WGS): Identifies genetic alterations. Transcriptomics (RNA-seq): Profiles gene expression. Proteomics (Mass Spectrometry): Quantifies protein abundance and modifications [65]. |
| Functional Assay Kits | Moves beyond correlative data to confirm the biological relevance and mechanistic role of a candidate biomarker. | Proliferation/Cell Viability Assays (e.g., CellTiter-Glo 3D): Measures drug response. Invasion/Migration Assays (e.g., Transwell): Assesses metastatic potential. CRISPR-Cas9 Knockout/Knockin Kits: Validates gene function. |
| AI/ML-Driven Data Analytics Platforms | Identifies complex, non-linear patterns in large, multi-dimensional datasets to predict clinical outcomes. | Predictive Modeling Software: Uses pre-clinical data to forecast patient response. Image Analysis Algorithms: Quantifies histopathological features from tissue sections. Integration Tools: Combines omics data with clinical variables. |
| Standardized Biomaterial Characterization Kits | Ensures accurate and reproducible assessment of material properties critical for biocompatibility and function. | Surface Characterization (e.g., ELISA-based kits): Quantifies protein adsorption. Degradation Profiling Assays: Measures resorption rates. Mechanical Testing Standards: Ensures consistent tensile/compressive strength data. |
The transition from pre-clinical discovery to clinical success hinges on the deliberate generation of high-quality, predictive data. As the biomaterials and drug development fields continue to expand, embracing an evidence-based framework is paramount. This involves a strategic shift from traditional models to human-relevant systems, integrating multi-omics data, insisting on functional validation, and leveraging advanced data analytics. By systematically implementing these strategies and rigorously documenting methodologies as outlined in this guide, researchers can significantly de-risk the translation pathway. This approach will ultimately accelerate the development of robust biomarkers and effective therapies, ensuring that pre-clinical promise translates into genuine clinical benefit for patients.
Leveraging AI and Machine Learning for Predictive Modeling and Data Analysis
The development of biomaterials has historically been characterized by a resource-intensive "trial-and-error" approach, often leading to substantial waste of manpower, time, materials, and finances [66]. The field now faces a paradigm shift with the integration of artificial intelligence (AI) and machine learning (ML), which provide data-driven methods to accelerate discovery and enhance predictive accuracy. This transition is foundational to evidence-based biomaterials research, a methodology that uses systematic approaches to translate research data into validated scientific evidence [5]. AI and ML models are uniquely positioned to address the high-dimensional complexity of biomaterial structure-property relationships, moving the field from intuition-driven experimentation to a quantitative, evidence-based discipline [67] [68] [66]. This guide objectively compares AI/ML methodologies, their performance in biomaterial applications, and the experimental protocols that underpin their predictive power, providing researchers with a framework for evaluating these tools within a rigorous evidence hierarchy.
AI and Predictive Analytics: Core Concepts and Comparative Framework
Understanding the distinction between AI and predictive analytics is crucial for selecting the appropriate tool for a research question.
Artificial Intelligence (AI) is technology that allows machines to perform tasks requiring human intelligence, such as analyzing data, recognizing patterns, or making decisions. It works by using algorithms, often powered by machine learning, that process large amounts of information, identify patterns, and apply those patterns to new situations [69]. In biomaterials, AI can handle work at a speed and scale that humans can't match.
Predictive Analytics is a data analysis method that uses historical data to forecast future outcomes. It is a subset of data analytics that sits alongside descriptive analytics (which looks at past results) and diagnostic analytics (which explains why they happened) [69]. Where descriptive and diagnostic analytics stop at the past, predictive analytics estimates what's likely to happen next.
These approaches are not mutually exclusive. Predictive analytics provides the framework for forecasting, while AI supplies advanced methods to improve accuracy or scale. They connect in the same workflow: predictive analytics generates a probability, and AI extends that forecast by drafting outreach or automating the next step [69]. A powerful emerging trend is the development of hybrid predictive AI/generative AI systems, where predictive AI can flag potentially unreliable generative AI outputs for human review, thereby increasing the overall reliability and autonomy of the system [70].
Table 1: Key Differences Between AI and Predictive Analytics
| Aspect | Artificial Intelligence (AI) | Predictive Analytics |
|---|---|---|
| Scope | Broad, covers pattern recognition, automation, content generation | Narrow, focuses on forecasting outcomes from historical data |
| Primary Function | Performing jobs that usually need human intelligence | Forecasting future outcomes like sales trends or customer churn |
| Data Handling | Can process both structured and unstructured data | Often starts with structured records like transactions or customer histories |
| Common Applications | AI-powered chatbots, fraud detection, generating personalized ad copy | Forecasting sales, predicting customer churn, projecting product demand |
Performance Comparison of Leading AI Models
For researchers selecting an AI model, key performance metrics include intelligence scores, speed, and context window capacity. The following data, synthesized from independent evaluations, provides a comparative overview of top 2025 models. Note that the "best" model depends heavily on the project's specific goals, budget, and infrastructure requirements [71].
Table 2: AI Model Performance Comparison (2025)
| Model | AAII Intelligence Index Score | Output Speed (tokens/sec) | End-to-End Time for 500 tokens (sec) | Max Context Window (tokens) |
|---|---|---|---|---|
| GPT-4o (OpenAI) | 85 | 60 | 10 | 8,192 |
| Claude 3.5 (Anthropic) | 80 | 45 | 18 | 100,000+ |
| LLaMA 3.1 (Meta) | 75 | 30 | 22 | 4,096 |
Key Takeaways:
- GPT-4o leads in speed and overall intelligence scores, ideal for businesses needing plug-and-play power and the snappiest user experience [71].
- Claude 3.5 excels in ethical AI and offers a massive context window, a game-changer for complex, multi-turn conversations or long document analysis, making it suitable for safety-critical applications [71].
- LLaMA 3.1 as an open-source model shines for customization and self-hosting, perfect for organizations with strict data privacy needs or custom infrastructure, though it requires more technical expertise [71].
Experimental Protocols for ML in Biomaterials Research
The application of ML in biomaterials follows distinct experimental workflows, primarily addressing two types of problems: the "forward problem" (property prediction) and the "inverse problem" (structure generation) [68]. The following protocol outlines a standard workflow for an ML-driven biomaterial discovery project.
Generalized Workflow for ML-Driven Biomaterial Discovery
ML-Driven Biomaterial Discovery Workflow
Detailed Methodology for Key Stages
1. Data Collection and Curation
- High-Throughput Experimentation (HTE): Generate large datasets using combinatorial libraries and automated synthesis. For example, Brocchini et al. studied the structure-function relationship in a combinatorial library of 112 distinct degradable polymers, performing 168 tests to study cell proliferation [67].
- Data Mining: Extract data from existing databases and scholarly publications. Challenges include polymer naming inconsistencies and high data sparsity for biomedical properties like in vivo degradation time and cytotoxicity [68].
- Data Preprocessing: Clean data, handle missing values, and perform feature engineering to create machine-readable representations of polymers (e.g., molecular graphs, BigSMILES strings) [68].
2. Problem Framing and ML Model Selection
- Forward Problem (Property Prediction): Given a polymer structure, predict specified properties of interest. This is a supervised learning task, ideal for screening candidate materials before synthesis [68].
- Inverse Problem (Structure Generation): Given target properties, predict polymeric structures that may demonstrate them. This expedites the materials design process beyond the limits of human intuition [68].
3. Model Training and Experimental Validation
- Training: Use curated datasets to train selected ML models (see Section 5.1). The model's performance is evaluated using technical metrics (e.g., precision, recall, AUC) and, crucially, business metrics like projected profit and savings to ensure operational value [70].
- Validation: Synthesize and characterize top candidate materials identified by the ML model to validate predictions. This step closes the loop and generates new, high-quality data to further refine the model [67] [68].
ML Applications in Biomaterials: Performance and Evidence Levels
The Scientist's Toolkit: Key ML Algorithms and Their Functions
Different ML algorithms are suited to specific tasks within biomaterial research. The table below details common algorithms and their applications.
Table 3: Essential ML Algorithms for Biomaterials Research
| Algorithm | Learning Type | Primary Function | Example Application in Biomaterials |
|---|---|---|---|
| K-Nearest Neighbour (KNN) | Supervised | Classification & Pattern Regression | Grouping biomaterials by similarity for property prediction [72]. |
| Random Forests | Supervised | Classification & Regression | Predicting scaffold degradation time from polymer structure [68]. |
| Decision Trees | Supervised | Classification & Regression | Identifying key structural features that drive cytotoxicity [66]. |
| Linear Regression | Supervised | Regression | Modeling the linear relationship between surface roughness and cell adhesion [66]. |
| Clustering (e.g., K-means) | Unsupervised | Discovering hidden patterns in data | Grouping unlabeled biomaterials data into conventional categories (metals, ceramics, polymers) [66]. |
| Reinforcement Learning | Reinforcement | Sequential decision-making | Optimizing 3D bioprinting parameters to maximize cell viability [66]. |
| Deep Neural Networks | Deep Learning | Complex pattern recognition from raw data | Analyzing medical images to predict the in vivo performance of an implant [66]. |
Evidence Hierarchy in AI-Driven Biomaterial Studies
The level of evidence in AI-based biomaterial research can be stratified, mirroring principles from evidence-based medicine [5]. The following diagram illustrates this hierarchy, from foundational data curation to the highest level of clinical impact.
Evidence Hierarchy for AI in Biomaterials
Comparative Performance of ML in Biomaterial Applications
ML has demonstrated significant efficacy across various biomaterial classes and applications. The following table summarizes documented performance.
Table 4: Performance of ML in Key Biomaterial Applications
| Application Area | Specific Task | ML Model(s) Used | Reported Performance / Outcome |
|---|---|---|---|
| Polymeric Biomaterials | Inverse design of new medical polymers | Random Forests, Gaussian Process Models | Identifies candidate structures, bypassing trial-and-error synthesis [68]. |
| Gene/Drug Delivery | Mapping structure-function of lipid nanoparticles (LNPs) | Data Mining coupled with ML | Accelerates discovery of optimal designs for drug encapsulation [67]. |
| Tissue Engineering Scaffolds | Predicting scaffold properties & optimizing design | Supervised Learning (Regression) | Enables data-driven design of 3D porous structures with desired properties [66]. |
| Antifouling Materials | Identifying physicochemical cues that reduce fouling | Data-driven discovery from high-throughput data | Maps complex structure-property relationships for faster discovery [67]. |
| Metallic Implants | Predicting effect of surface topography on cell response | Pattern Recognition | Shows that increasing surface roughness can improve osteoblast spreading [67]. |
| Biomechanics | Regulating tissue growth/remodelling with mechanical stimuli | Various ML models for in-silico modelling | Unveils complex relationships among engineering, biomaterials, and biomedicine factors [72]. |
The integration of AI and ML into biomaterials science marks a transformative shift from a trial-and-error paradigm to a quantitative, evidence-based discipline. As the field matures, the combination of predictive and generative AI in hybrid systems, alongside the adoption of disciplined business practices like bizML for project valuation, promises to further increase the success rate of deployments [70]. For researchers, the key lies in selecting the appropriate tool—be it a high-performance proprietary model like GPT-4o for rapid analysis, an open-source platform like LLaMA 3.1 for secure data handling, or a specific ML algorithm for property prediction—based on the problem's specific requirements, available data, and the desired level of evidence. By leveraging these powerful tools within a rigorous methodological framework, the biomaterials community can dramatically accelerate the discovery and development of next-generation medical products.
Addressing Biocompatibility Challenges and Immune Response Evaluation
The clinical success of any implantable medical product is fundamentally governed by its biocompatibility, which is determined by the nature and extent of the immune response it elicits upon implantation. The paradigm in biomaterials science has progressively shifted from developing passive, bioinert materials that minimally interact with the host toward engineering bioactive and smart immunomodulatory materials that actively direct favorable immune responses for improved integration and tissue regeneration [73] [74]. This evolution necessitates robust, standardized evaluation methodologies to objectively compare the performance of emerging biomaterials against established alternatives.
Framing this evaluation within the context of evidence-based biomaterials research (EBBR) is crucial for translating promising in vitro results into clinically viable products [4]. EBBR employs systematic reviews and meta-analyses of existing research data to generate validated scientific evidence, creating a hierarchy of evidence that helps distinguish experimental artifacts from physiologically relevant outcomes. This review will objectively compare the immune responses elicited by major biomaterial classes, provide detailed experimental protocols for their evaluation, and present a toolkit for researchers in the field.
Comparative Analysis of Biomaterial Classes and Immune Responses
Different biomaterial classes interact with the immune system in distinct ways, leading to varied outcomes in terms of inflammation, fibrosis, and tissue integration. The following section compares the immune performance of synthetic polymers, metals, ceramics, and natural biological scaffolds.
Table 1: Comparative Immune Response Profiles of Major Biomaterial Classes
| Biomaterial Class | Key Immune Interactions | Macrophage Polarization Tendency | Typical Fibrosis Response | Representative Experimental Data |
|---|---|---|---|---|
| Synthetic Polymers (e.g., PLGA, PCL) | Protein adsorption; Foreign Body Response (FBR) leading to giant cell formation [73] [75] | Predominantly pro-inflammatory M1, potentially transitioning to M2 with modification [73] | High risk of fibrous capsule formation [73] | ~80% of implants show fibrotic encapsulation >50μm thick [73] |
| Metals & Alloys (e.g., Titanium, Vitallium) | Bioinertness; Ion release can modulate immune cells; Surface topography critical [73] [76] | M1 with smooth surfaces; M2-promoting with specific nanostructuring [73] | Low with optimal surface bioactivity [73] | Surface nanopatterning reduced inflammation markers by >60% in vivo [73] |
| Bioceramics & Bioactive Glasses | Direct ionic exchange; Osteoimmunomodulation [77] [76] | Can promote M2 polarization via ion release (e.g., Ca²âº, Siâ´âº) [77] | Low to moderate, integration common in bone [76] | Strontium-doped ceramics increased IL-10 (M2 marker) by 3.5-fold vs. controls [77] |
| Natural ECM Bioscaffolds | Dynamic interaction with DAMPs; MBV as signaling entities [75] | Strongly promotes transition to pro-regenerative M2 phenotype [75] | Constructive remodeling instead of fibrotic encapsulation [75] | ~90% constructive remodeling vs. <20% for synthetic mesh in musculofascial repair [75] |
Key Insights from Comparative Analysis
The data reveals a clear spectrum of immune compatibility. Synthetic polymers, while versatile and mechanically tunable, consistently trigger a pronounced foreign body response, often culminating in fibrous encapsulation that isolates the implant and can lead to failure [73]. Their performance is highly dependent on specific formulations and surface modifications.
Metals and ceramics demonstrate more favorable profiles, particularly in orthopedic applications. Their bioactivity—the ability to form a direct bond with bone tissue—is closely linked to their immunomodulatory potential. For instance, the release of specific ions from bioactive glasses can directly shift macrophage polarization toward the pro-healing M2 phenotype, creating a microenvironment conducive to osteogenesis [77] [76].
The most favorable immune outcomes are observed with decellularized extracellular matrix (ECM) bioscaffolds. Unlike synthetic materials that often lead to chronic inflammation, well-prepared ECM scaffolds interact dynamically with host immune cells. They preserve native structural and biochemical cues, such as matrix-bound nanovesicles (MBVs), which promote a controlled inflammatory phase that rapidly resolves and transitions into a pro-regenerative, anti-inflammatory state supporting constructive tissue remodeling [75]. The host recognizes these scaffolds not as foreign, but as a natural template for repair.
Experimental Protocols for Immune Response Evaluation
A tiered methodology, progressing from in vitro screening to in vivo validation, is essential for a comprehensive evidence-based evaluation of biomaterial immune compatibility.
In Vitro Macrophage Polarization Assay
Purpose: To quantitatively assess the intrinsic propensity of a biomaterial to drive pro-inflammatory (M1) or pro-healing (M2) macrophage polarization.
Detailed Protocol:
- Macrophage Culture: Use human monocyte cell lines (e.g., THP-1) differentiated with PMA (Phorbol 12-myristate 13-acetate) or primary human monocyte-derived macrophages (MDMs).
- Material Exposure: Seed cells and expose to biomaterial extracts (ISO 10993-12) or directly culture on 3D material scaffolds. Include controls: LPS (Lipopolysaccharide) + IFN-γ for M1 and IL-4 for M2 polarization.
- Analysis (72 hours post-exposure):
Data Interpretation: A favorable immunomodulatory material will show a significant upregulation of M2 markers and a concomitant downregulation of M1 markers compared to a negative control (e.g., a material known to elicit a strong FBR).
In Vivo Subcutaneous Implantation Model
Purpose: To evaluate the full spectrum of the foreign body response, including chronic inflammation and fibrosis, within a physiologically relevant tissue microenvironment.
Detailed Protocol:
- Implantation: Following ethical approval, implant material samples (e.g., 5x5mm discs) into subcutaneous pockets of a rodent model (e.g., Sprague-Dawley rats). Use inert materials (e.g., medical-grade silicone) and pro-remodeling materials (e.g., ECM scaffold) as controls.
- Explanation and Analysis: Harvest implants at multiple time points (e.g., 1, 2, and 4 weeks).
- Histology: Process for H&E staining for general morphology and Masson's Trichrome for collagen deposition/fibrosis.
- Immunohistochemistry/Immunofluorescence: Stain for cell-specific markers:
- Neutrophils: MPO (Myeloperoxidase) - early inflammation.
- Macrophages: F4/80 (pan-macrophage), iNOS (M1), CD206 (M2).
- T cells: CD3âº.
- Myofibroblasts: α-SMA (Alpha-Smooth Muscle Actin) - fibrosis indicator.
- Capsule Thickness: Measure fibrous capsule thickness around the implant from multiple Trichrome-stained sections [73] [75].
Data Interpretation: A superior biocompatibility profile is indicated by a thin, vascularized tissue layer with a high density of M2 macrophages and minimal fibrous capsule formation at the 4-week endpoint, demonstrating constructive remodeling rather than isolation.
Diagram 1: The in vivo immune response to biomaterial implantation progresses through acute inflammation, a critical decision point in macrophage polarization, and culminates in either fibrotic encapsulation (failure) or constructive tissue remodeling (success).
The Scientist's Toolkit: Key Research Reagent Solutions
A standardized set of reagents and tools is fundamental for generating comparable and reproducible data in biomaterial-immune response research.
Table 2: Essential Research Reagents for Evaluating Biomaterial Immune Compatibility
| Reagent/Tool Category | Specific Examples | Function in Evaluation |
|---|---|---|
| Immune Cell Lines & Primary Cells | THP-1 (human monocytes), RAW 264.7 (mouse macrophages), Primary human MDMs | Provide a renewable and physiologically relevant cell source for in vitro polarization assays [77] [73]. |
| Polarization Inducers & Inhibitors | LPS (Lipopolysaccharide), IFN-γ (Interferon-gamma), IL-4 (Interleukin-4), IL-13, CSF1R inhibitors | Positive and negative controls for directing M1/M2 polarization; used to test specific immunomodulatory hypotheses [73]. |
| Antibodies for Flow Cytometry/IF | Anti-CD68 (pan-macrophage), anti-iNOS (M1), anti-CD206 (MMR, M2), anti-CD3 (T-cells), anti-α-SMA (myofibroblasts) | Enable identification and quantification of specific immune cell populations and phenotypes in vitro and in tissue sections [77] [75]. |
| Cytokine Detection Kits | ELISA or Multiplex Luminex kits for TNF-α, IL-1β, IL-6, IL-12 (M1), IL-10, TGF-β (M2) | Quantify the secretory profile of immune cells, providing a functional readout of the inflammatory state [77]. |
| Decellularized ECM Controls | Commercially available urinary bladder matrix (UBM) or small intestinal submucosa (SIS) ECM | Serve as a positive control for pro-remodeling immune responses in comparative studies [75]. |
Advanced Models and Future Perspectives
The field is rapidly advancing beyond traditional models. Microfluidic Immune-System-on-a-Chip (ISOC) platforms are emerging as powerful tools that recapitulate human immune-tumor interactions and dynamic immune cell activity, offering a more predictive model for screening immunomodulatory biomaterials, particularly for cancer and advanced therapeutic applications [78]. Furthermore, the development of stimuli-responsive "smart" biomaterials that release immunomodulatory cargo (e.g., anti-inflammatory cytokines) in response to environmental cues like pH or enzymes represents the next frontier in precision immune engineering [79] [74].
Diagram 2: Intracellular signaling pathways driving macrophage polarization. Material-derived signals initiate distinct pathways: pro-inflammatory M1 via NF-κB/MAPK or pro-regenerative M2 via JAK-STAT6. Ions released from bioceramics can promote the M2 pathway.
A rigorous, evidence-based approach to evaluating biocompatibility and immune responses is non-negotiable for the development of next-generation medical devices and implants. The comparative data clearly indicates that the biological properties of the material—particularly its ability to guide macrophage polarization toward a pro-regenerative M2 phenotype—are as critical as its structural and mechanical properties. By employing the standardized experimental protocols and research tools outlined herein, researchers can generate high-quality, comparable data. This will accelerate the translation of advanced immunomodulatory biomaterials from the laboratory to the clinic, ultimately leading to implants that are not just tolerated by the body, but actively harness the immune system to achieve better patient outcomes.
Optimizing High-Throughput Screening and Material Informatics
The translation of biomaterials from basic research to commercial medical products follows a structured roadmap, generating different levels of scientific evidence at each stage. Basic research, driven by curiosity and hypotheses, explores unknown knowledge with no direct path to commercialization. Applied research targets potential applications, producing prototypes and processes. The critical transition to product development requires rigorous non-clinical evaluation (bench tests, biocompatibility) and clinical evaluation under quality management systems, culminating in post-market surveillance and real-world evidence [18]. Within this framework, High-Throughput Screening (HTS) and Materials Informatics (MI) represent powerful, complementary methodologies that enhance the efficiency and robustness of evidence generation during the early discovery and optimization phases. This guide objectively compares their performance in accelerating biomaterials innovation.
Experimental Protocols and Methodologies
High-Throughput Screening (HTS) Protocol
HTS uses automated equipment to rapidly test thousands to millions of samples for biological activity. The following protocol details a typical qHTS campaign, which tests compounds at multiple concentrations to generate concentration-response curves [80] [81].
- Step 1: Assay Design and Miniaturization. A biochemical or cellular assay is designed to model the target biological activity. The assay is miniaturized into microtiter plates (96-, 384-, or 1536-well formats) to reduce reagent costs and increase throughput [80].
- Step 2: Library Management and Reagent Dispensing. A library of small molecules, natural product extracts, or other substances is prepared in dimethyl sulfoxide (DMSO) stocks. Robotic liquid handlers transfer nanoliter to microliter volumes of compounds and assay reagents into the plates [80].
- Step 3: Quantitative Screening Incubation. The assay plates are incubated under controlled conditions. In qHTS, this is performed across a series of plates, each containing a different concentration of the test compounds, typically spanning four to eight orders of magnitude [81].
- Step 4: Signal Detection and Primary Analysis. Post-incubation, a signal (e.g., absorbance, fluorescence, luminescence) is measured using automated plate readers. Raw data is processed to calculate a response value for each well, normalized to positive (no inhibition) and negative (full inhibition) controls [80].
- Step 5: Curve Fitting and Hit Identification. For each compound, response data across all concentrations is fitted to a nonlinear model, most commonly the Hill equation (Equation 1), to estimate key parameters: AC~50~ (potency), E~max~ (efficacy), and h (Hill slope) [81]. Compounds meeting predefined activity criteria (e.g., AC~50~ < 10 µM, E~max~ > 80%) are classified as "hits."
Equation 1: Hill Equation
Where R~i~ is the measured response at concentration C~i~, E~0~ is the baseline response, and E~∞~ is the maximal response [81].
Materials Informatics (MI) Protocol
Materials Informatics applies data-centric approaches, leveraging machine learning (ML) to discover and optimize materials. The following workflow is common in data-driven materials discovery [82] [83].
- Step 1: Data Curation and Integration. Structured and unstructured data is aggregated from internal experiments (e.g., HTS), computational simulations (e.g., molecular dynamics), and external repositories. This includes chemical structures, processing conditions, and performance properties [83].
- Step 2: Feature Engineering and Representation. Materials are converted into a computer-readable format using descriptors or fingerprints (e.g., molecular weight, atomic coordinates, chemical bonds). Domain knowledge is critical for selecting meaningful features from high-dimensional, sparse data [82].
- Step 3: Machine Learning Model Training. A dataset of known materials and their properties is used to train an ML model (e.g., random forest, neural networks). The model learns the complex relationships between a material's features and its target properties [82] [83].
- Step 4: Predictive Screening and Inverse Design. The trained model screens a vast virtual library of candidate materials, predicting their properties and performance. In the idealized "inverse design" approach, the model generates new material structures that satisfy a set of desired properties [82].
- Step 5: Experimental Validation and Active Learning. A shortlist of top-predicted candidates is synthesized and tested experimentally. The results from these validation experiments are fed back into the dataset to refine the ML model in an iterative active learning loop, reducing the number of experiments required [82].
Performance Comparison and Experimental Data
The table below summarizes a quantitative comparison of HTS and MI based on key performance metrics, supported by experimental data and case studies.
Table 1: Performance Comparison of HTS and Materials Informatics
| Metric | High-Throughput Screening (HTS) | Materials Informatics (MI) |
|---|---|---|
| Throughput | High: 10^3^ - 10^6^ compounds screened in parallel [80]. | Ultra-High: Virtual screening of 10^6^ - 10^12^ candidates computationally [83]. |
| Hit Rate | Low to Moderate: Traditional HTS hit rates can be as low as 0.01% for challenging targets like protein-protein interactions [80]. | Higher and More Focused: AI pre-screening leads to enriched candidate lists, significantly improving experimental hit rates [83]. |
| Cycle Time | Months: A single qHTS campaign can take several months from setup to hit confirmation [80]. | Weeks to Months: Initial model setup and training can be rapid, with iterative design-make-test cycles taking weeks [82]. |
| Data Output | Concentration-response curves for thousands of compounds (HEQN parameters: AC~50~, E~max~, h) [81]. | Predictive models and property predictions for vast chemical spaces (e.g., conductivity, stability) [83]. |
| Cost Implications | High capital investment in robotics and reagents. Cost per data point is low, but total cost is high due to massive scale [80]. | High initial data infrastructure and AI expertise cost. Reduces long-term R&D costs by minimizing failed experiments [84] [83]. |
| Key Limitations | - High false positive/negative rates in single-concentration HTS [80].- Parameter estimate variability in qHTS if asymptotes are not defined [81].- Limited to available compound libraries. | - Dependence on quality/quantity of training data [82].- Sparse, high-dimensional, and noisy data challenges [82].- High implementation cost for SMEs [83]. |
A case study in battery innovation demonstrates the power of MI. An EV manufacturer used AI-driven predictive modeling and deep tensor learning to screen thousands of electrode materials. This approach identified a high-performing, cobalt-free alternative, reducing the discovery cycle from 4 years to under 18 months and lowering R&D costs by 30% by reducing trial-and-error experimentation [83].
Workflow Visualization
The following diagrams illustrate the core workflows for HTS and MI, highlighting their iterative, evidence-generating nature.
Diagram 1: The qHTS workflow generates quantitative dose-response data for hit identification.
Diagram 2: The MI workflow uses an active learning loop to continuously improve predictive models.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Research Reagent Solutions for HTS and MI
| Item | Function in Research |
|---|---|
| Microtiter Plates (384-/1536-well) | The standard platform for assay miniaturization in HTS, enabling parallel processing of thousands of reactions with minimal reagent volumes [80]. |
| HTS Compound Libraries | Curated collections of hundreds of thousands of small molecules or biologics used for screening against a biological target to find initial hits [80]. |
| Biochemical/Cell-Based Assay Kits | Standardized reagents for measuring specific biological activities (e.g., kinase activity, cell viability). Critical for ensuring reproducibility and quality in HTS [80]. |
| Laboratory Automation Robotics | Automated liquid handlers, plate washers, and incubators that enable the rapid and precise execution of HTS protocols without manual intervention [80]. |
| Materials Data Cloud Platforms | Scalable informatics platforms (e.g., Citrine Informatics, Schrödinger) for storing, managing, and analyzing large-scale materials data [82] [83]. |
| Simulation & Modeling Software | Computational tools (e.g., for Density Functional Theory - DFT) used to generate high-quality synthetic data for training MI models where experimental data is scarce [83]. |
Validation and Comparison: Assessing Biomaterial Safety, Efficacy, and Performance
The translation of biomaterials from laboratory research to clinical application requires rigorous validation across a structured framework of standards. These frameworks, including Good Laboratory Practice (GLP), current Good Manufacturing Practice (cGMP), and ISO 10993 for biological evaluation, establish a continuum of evidence generation from non-clinical safety to clinical performance. Understanding the distinct roles, requirements, and interrelationships of these frameworks is fundamental for researchers, scientists, and drug development professionals aiming to navigate the complex pathway of biomaterial validation.
This guide provides a comparative analysis of these key validation frameworks, contextualized within a broader thesis on evidence levels in biomaterial research. It examines how each framework contributes to building a comprehensive safety and efficacy profile, supported by experimental data and standardized protocols.
Framework Definitions and Regulatory Context
Good Laboratory Practice (GLP)
Good Laboratory Practice (GLP) is a quality system governing the non-clinical safety testing of products in laboratory settings. Originating from US FDA regulations in the late 1970s following cases of data falsification, GLP principles ensure the integrity and reliability of safety data submitted to regulatory authorities [85]. GLP applies specifically to non-clinical health and environmental safety studies, providing a framework for study planning, performance, monitoring, recording, reporting, and archiving [86] [85]. A cornerstone of GLP is the requirement for a Quality Assurance (QA) programme that independently verifies that all written procedures, including the study protocol, are followed and that the final report accurately represents the results [85].
Current Good Manufacturing Practice (cGMP)
Current Good Manufacturing Practice (cGMP) comprises regulations ensuring that manufactured products consistently meet quality standards appropriate to their intended use [86]. Unlike GLP's focus on research data integrity, cGMP applies to the commercial manufacturing process itself, emphasizing controlled production environments, detailed standard operating procedures (SOPs), and comprehensive documentation [86]. The "current" designation underscores the expectation that manufacturers employ up-to-date technologies and systems. cGMP controls are often summarized as the "Five Ps": People, Products, Processes, Procedures, and Premises, all requiring stringent quality management [86].
ISO 10993 (Biological Evaluation of Medical Devices)
The ISO 10993 series of standards, specifically ISO 10993-1, outlines principles for the biological evaluation of medical devices within a risk management process [87]. The 2025 revision represents a significant evolution from a prescriptive "checklist" approach to a risk-based framework fully integrated with ISO 14971 (Risk Management for Medical Devices) [87] [88]. The primary aim is protecting humans from potential biological risks arising from medical device use by evaluating factors such as device nature, material composition, and contact duration with the body [87] [89]. The framework requires a structured biological evaluation process that includes hazard identification, risk estimation, and risk control throughout the device lifecycle [87].
Comparative Analysis of Frameworks
The table below summarizes the core objectives, regulatory focus, and primary application stages for GLP, cGMP, and ISO 10993.
Table 1: Core Characteristics of Validation Frameworks
| Framework | Primary Objective & Focus | Regulatory/Standardization Context | Stage in Product Lifecycle |
|---|---|---|---|
| GLP | Ensure data integrity and reliability of non-clinical safety studies [86] [85]. | OECD Principles, national law in many countries; FDA 21 CFR Part 58 [85]. | Pre-market R&D and safety testing [86]. |
| cGMP | Ensure consistent production quality and patient safety of commercial products [86]. | FDA regulations (e.g., 21 CFR Parts 210 & 211); international equivalents [86]. | Commercial manufacturing and post-market quality control [86]. |
| ISO 10993-1 | Manage biological risk from medical devices; protect humans from biological hazards [87] [89]. | International standard; recognized by regulatory bodies globally (e.g., FDA, EU MDR) [87] [88]. | Device design, development, and post-market surveillance [87]. |
Interrelationship and Workflow
These frameworks do not operate in isolation but form a connected pathway from discovery to market. The following diagram visualizes this logical progression and the primary focus of each framework.
Diagram: The sequential relationship and primary focus of key validation frameworks in the medical product lifecycle.
Experimental Protocols and Methodologies
GLP-Compliant Safety Study Protocol
A typical GLP-compliant safety study for a biomaterial involves a meticulously documented and controlled process.
- Test Facility and Personnel: The study must be conducted in a facility with a defined organizational structure. The Study Director bears single-point responsibility for the overall study conduct and final report [85]. All personnel must have the education, training, and experience to perform their assigned functions.
- Quality Assurance: The independent QA unit monitors the entire study, verifying that the protocol is followed and the report accurately reflects the raw data [85].
- Study Plan (Protocol): A detailed, pre-approved protocol defines the study's objective, materials, methods, and scheduling. It specifies the test system, dose levels, route of administration, and type/frequency of analyses [85].
- Test System and Test Item: The test system (e.g., animals, cell lines) must be well-characterized. The test item (the biomaterial) must be appropriately characterized and stored, with its stability and concentration verified [85].
- Standard Operating Procedures (SOPs): All critical operations follow written SOPs to ensure consistency and data quality [85].
- Data Generation and Recording: All original observations are recorded directly, promptly, and legibly, with any changes tracked. This ensures full data traceability and integrity [85].
- Final Report: The final report, signed by the Study Director, includes a complete description of methods, results, a discussion, and conclusions, and it must exactly reflect the raw data [85]. All raw data, samples, and the final report are archived for a defined period.
ISO 10993-1:2025 Biological Evaluation Process
The updated ISO 10993-1:2025 mandates a risk-based biological evaluation integrated within a quality management system. The workflow below outlines this iterative process.
Diagram: The iterative biological evaluation process for medical devices as per ISO 10993-1:2025 risk management framework [87].
Key methodological considerations per ISO 10993-1:2025 include:
- Biological Evaluation Plan (BEP): The BEP is foundational, defining the device, its intended use, and reasonably foreseeable misuse [87]. It must document the biological effects to be addressed, acceptance criteria, and the rationale for all decisions.
- Device Categorization: Contact duration is calculated conservatively. Each day of exposure counts as one full day, regardless of contact time within that day. The categories are limited (<24h), prolonged (24h-30d), and long-term (>30d) [87] [88]. Furthermore, device categorization now focuses solely on the nature of body contact (intact skin, mucosa, breached surfaces/tissues, blood) rather than the "external communicating" device type [88].
- Chemical Characterization: A thorough identification and quantification of leachable substances is critical for toxicological risk assessment, per ISO 10993-18 [87] [89] [88].
- Risk Estimation and Evaluation: This is a core update in the 2025 version. It requires estimating biological risk based on the severity of harm and probability of its occurrence, directly aligning with ISO 14971 methodology [87].
- Testing and Justification: Testing for biological effects (cytotoxicity, sensitization, etc.) is not automatic but is driven by the risk assessment. The standard emphasizes avoiding animal testing via the 3R principles (Replace, Reduce, Refine) when possible [88].
Essential Research Reagents and Materials
The table below details key reagents and materials essential for conducting experiments within these validation frameworks.
Table 2: Key Research Reagent Solutions for Biomaterial Validation
| Reagent / Material | Primary Function in Validation | Application Context |
|---|---|---|
| Reference Materials | Serve as benchmarks for calibration and method validation; ensure data comparability across studies and labs. | Used in GLP studies for apparatus calibration and in chemical characterization per ISO 10993-18. |
| Cell Lines (e.g., L929) | In vitro assessment of cytotoxicity (ISO 10993-5); provide a sensitive indicator of a material's potential to cause cell damage or death. | Initial biocompatibility screening of device extracts in a biological evaluation plan. |
| Test/Reference Items | The well-characterized biomaterial or device itself; the subject of the safety or efficacy study. | The central item in any GLP safety study or ISO 10993 biological evaluation. |
| Culture Media & Reagents | Support the growth and maintenance of test systems (cells, tissues); quality is critical for reproducible results. | Used in cytotoxicity, sensitization, and other in vitro tests within a biological evaluation. |
| Extraction Solvents | Extract leachable and semi-volatile compounds from a device for chemical characterization and toxicological risk assessment. | Applied per ISO 10993-12 to simulate the release of substances under clinical-use conditions. |
| Histological Stains | Enable microscopic visualization and scoring of tissue response in implantation studies (e.g., H&E staining). | Critical for evaluating local effects after implantation as part of ISO 10993-6 evaluation. |
The journey of a biomaterial from a research concept to a clinically approved product demands adherence to a multi-stage, interconnected validation pathway. GLP provides the foundation of reliable non-clinical safety data, cGMP ensures the quality and consistency of its manufacture, and ISO 10993 delivers a structured, risk-based framework for evaluating its biological safety. The 2025 update to ISO 10993-1, with its deepened integration of risk management principles, marks a significant shift from a prescriptive to a knowledge-driven approach. For researchers and developers, a clear understanding of the distinct yet complementary roles of these frameworks is not merely a regulatory hurdle but a fundamental component of rigorous scientific practice and successful clinical translation.
The selection of an appropriate biomaterial is a fundamental determinant of success in medical applications, from everyday medical devices to cutting-edge regenerative medicine. The field has evolved significantly from using first-generation, bio-inert materials to a new era of third-generation materials designed to actively direct specific biological responses such as tissue regeneration [90]. This evolution demands a rigorous, evidence-based framework for material selection. This guide provides a comparative analysis of four principal biomaterial classes—polymers, ceramics, metals, and hydrogels—contextualized within a hierarchy of scientific evidence. It aims to equip researchers and developers with a structured approach to evaluating biomaterial performance, moving from in vitro characterization to pre-clinical and clinical validation. By integrating quantitative data, experimental protocols, and a clear evidence-level framework, this analysis supports informed decision-making in therapeutic and device development.
Biomaterial Classes and Key Characteristics
The distinct physical, chemical, and biological properties of each biomaterial class make them suitable for different clinical applications. The global biomaterials market, valued at $134.31 billion in 2024 and projected to grow rapidly, reflects the critical importance of these materials in healthcare [91]. The following section provides a detailed, evidence-based comparison of the four main classes.
Table 1: Comparative Overview of Major Biomaterial Classes
| Biomaterial Class | Key Subtypes & Examples | Primary Advantages | Primary Limitations & Evidence Gaps | Common Clinical Applications |
|---|---|---|---|---|
| Polymers | Synthetic: PLA, PGA, PCL, PEG, PU [90]Natural: Collagen, Chitosan, Hyaluronic Acid [90] | Versatile and tunable properties; biodegradable; can be engineered for controlled drug release [90]. | Potential poor biocompatibility (synthetics); batch-to-batch variability (naturals); long-term biodegradation products require more clinical safety data [90]. | Sutures, vascular grafts, drug delivery systems, tissue engineering scaffolds [90]. |
| Ceramics | Bioceramics (e.g., Hydroxyapatite), Porous and Non-porous Ceramics [91] | High compressive strength; excellent biocompatibility and osteoconductivity; bioresorbable [91]. | Brittleness; low tensile strength and fracture toughness; limited evidence for load-bearing bone repair [91]. | Dental implants, bone graft substitutes, coatings for metallic orthopedic implants [91]. |
| Metals | Stainless Steel, Titanium Alloys, Cobalt-Chromium Alloys, Magnesium Alloys [91] | Superior tensile strength, fatigue resistance, and durability. | High stiffness leading to stress-shielding; potential for corrosion and metal ion release; not biodegradable (except Mg alloys, which need more clinical study) [91]. | Fracture fixation plates, joint replacements, stents, dental roots [91]. |
| Hydrogels | Natural-based: Gelatin, Chitosan, Alginate [92]Synthetic: PVA, PAM, PNIPAAm [92] | High water content; tissue-like mechanical compliance; inherent biocompatibility; tunable porosity for drug delivery [93]. | Often mechanically weak; complex and sometimes irreproducible fabrication [93]; long-term stability in vivo requires further validation. | Wound dressings, drug delivery, tissue engineering scaffolds, contact lenses [93]. |
Table 2: Quantitative Performance Data for Biomaterial Classes
| Property | Polymers | Ceramics | Metals | Hydrogels |
|---|---|---|---|---|
| Young's Modulus | 0.1 MPa - 5 GPa [90] | 50 - 500 GPa [91] | 100 - 230 GPa [91] | 0.1 - 100 kPa [92] |
| Tensile Strength | Low - High (Varies widely by type) [90] | Very Low (Brittle) | Very High | Very Low |
| Biodegradability | Tunable (days to years) [90] | Slow (months to years) | Non-degradable (except Mg alloys) | Tunable (hours to months) [93] |
| Bioactivity | Can be engineered; natural polymers are inherently bioactive [90] | Highly osteoconductive | Bio-inert (integration relies on surface modifications) | Can be engineered to be bioactive [93] |
A key trend in biomaterials research is the development of composite and hybrid systems that combine classes to overcome individual limitations. A prominent example is the creation of piezoelectric hydrogels, which integrate energy-harvesting piezoelectric materials (often ceramics or polymers) into a soft, compliant hydrogel matrix. This combination yields a self-powered, biomimetic material capable of providing electrical stimulation for enhanced wound healing or nerve regeneration [94]. Similarly, polymer-ceramic composites are widely used in orthopedics to create scaffolds with improved bone-binding ability and mechanical resilience.
Evidence Framework in Biomaterials Research
The concept of Evidence-Based Biomaterials Research (EBBR) has been proposed to systematically translate research data into validated scientific evidence, mirroring the framework of Evidence-Based Medicine [4]. This is crucial for navigating the vast and complex landscape of biomaterials literature and for making informed decisions from the lab to the clinic.
The hierarchy of evidence, adapted for biomaterials science, is visualized below. This framework helps contextualize the strength of conclusions drawn from different types of studies, with systematic reviews of randomized controlled trials (RCTs) representing the highest level of evidence for clinical outcomes [9].
The foundation of the pyramid consists of in vitro and computational studies, which provide essential data on material properties and initial biocompatibility. For instance, a study screening 180 bioinspired hydrogels for adhesive strength is a Level 5 study that generates hypotheses for future in vivo testing [95]. As research moves up the pyramid, the biological complexity and clinical relevance increase, but so do the cost and time required. Systematic reviews and meta-analyses, which statistically synthesize findings from multiple high-quality studies, offer the most reliable and generalizable conclusions for clinical decision-making [4].
Experimental Analysis and Workflows
A standardized approach to biomaterial evaluation is critical for generating comparable and reliable data. The following workflow outlines a generalized protocol for the in vitro and in vivo assessment of a new biomaterial, such as a hydrogel for bone regeneration.
Detailed Experimental Protocols
Protocol 1: Synthesis of Data-Driven Adhesive Hydrogels
This protocol, based on a high-impact study, demonstrates a modern approach integrating data mining and machine learning for material design [95].
- Objective: To design and synthesize super-adhesive hydrogels through a data-driven, bioinspired approach.
- Materials:
- Functional Monomers: Six monomers representing hydrophobic, nucleophilic, acidic, cationic, amide, and aromatic classes of amino acids.
- Crosslinker: e.g., Methylene bisacrylamide.
- Solvent: Dimethyl sulfoxide (DMSO).
- Initiator: e.g., Ammonium persulfate (APS).
- Catalyst: e.g., N,N,N',N'-Tetramethylethylenediamine (TEMED).
- Method:
- Data Mining: Mine adhesive protein databases (e.g., NCBI) to identify consensus sequences from various species. Encode these sequences using a functional class descriptor (grouping the 20 amino acids into 6 classes).
- Formulation Derivation: Calculate the relative composition (Ï•i) of each functional class for the top species. These compositions serve as the formulations for the hydrogels.
- Ideal Random Copolymerization: Synthesize hydrogels via one-pot free-radical copolymerization in DMSO, which ensures near-unity reactivity ratios for statistically controlled sequences.
- Post-processing: After polymerization, exchange the solvent from DMSO to normal saline (0.154 M NaCl) to form the final hydrogel.
- Characterization:
- Adhesive Strength (Fa): Measure using a tack test on a glass substrate in normal saline, with a defined loading force (e.g., 10 N) and contact time (e.g., 10 s) [95].
- Swelling Ratio: Determine the equilibrium swelling ratio in saline.
- Rheology: Perform oscillatory shear measurements to determine storage (G') and loss (G'') moduli.
Protocol 2: Evaluating Immunomodulation for Bone Regeneration
This protocol assesses a biomaterial's interaction with the immune system, a critical factor for successful integration and regeneration [77].
- Objective: To evaluate the capacity of a biomaterial (e.g., a ceramic bone graft or an immunomodulatory hydrogel) to modulate macrophage polarization and subsequent osteogenic outcomes.
- Materials:
- Macrophage Cell Line: e.g., RAW 264.7 or primary bone marrow-derived macrophages (BMDMs).
- Culture Medium: DMEM/F12 supplemented with FBS and antibiotics.
- Polarizing Cytokines: LPS and IFN-γ for M1 polarization; IL-4 for M2 polarization.
- Conditioned Media Protocol: Culture macrophages with the test biomaterial extract or by direct contact, then collect the conditioned media.
- Osteoprogenitor Cells: e.g., MC3T3-E1 pre-osteoblasts or human Mesenchymal Stem Cells (hMSCs).
- Method:
- Macrophage Culture and Polarization: Culture macrophages and stimulate them toward an M1 phenotype. Co-culture the polarized macrophages with the test biomaterial.
- Conditioned Media Transfer: After a set period (e.g., 48-72 hours), collect the conditioned media from the macrophage-biomaterial cultures.
- Osteogenic Assay: Culture osteoprogenitor cells in the conditioned media under osteogenic conditions. Run appropriate controls (e.g., conditioned media from M1 macrophages without biomaterial).
- Analysis:
- Macrophage Phenotype: Quantify M1 (e.g., iNOS, TNF-α) and M2 (e.g., ARG1, CD206) markers via qPCR or immunofluorescence.
- Osteogenic Differentiation: Measure alkaline phosphatase (ALP) activity, perform Alizarin Red S staining for mineralization, and analyze osteogenic gene expression (e.g., Runx2, OPN) in the progenitor cells.
- In Vivo Correlative Model: Implant the biomaterial in a critical-sized calvarial defect in a rat. After 4-8 weeks, analyze the explants via histology (H&E, staining for M1/M2 macrophages) and micro-CT to quantify new bone volume.
The complex signaling cascades involved in bone immunomodulation are summarized below.
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Biomaterials Research
| Reagent / Material | Function in Research | Specific Examples & Notes |
|---|---|---|
| Functional Monomers | Building blocks for creating polymeric and hydrogel networks with specific chemical properties. | Six-class monomers for bioinspired adhesives [95]; Methacrylated gelatin (GelMA) for photopolymerizable hydrogels. |
| Crosslinkers | To create stable 3D networks in polymers and hydrogels, determining mechanical strength and degradation. | Methylene bisacrylamide (chemical); Genipin (for natural polymers); UV light for photo-crosslinking. |
| Piezoelectric Fillers | To impart energy-harvesting capability to composites for self-powered electrical stimulation. | Barium Titanate (BTO) ceramics; Poly(vinylidene fluoride) (PVDF) polymers [94]. |
| Cell Culture Assays | To evaluate cytocompatibility and biofunctionality according to international standards (ISO 10993-5). | AlamarBlue (cell viability); Alkaline Phosphatase (ALP) kit (early osteogenesis); Alizarin Red S (mineralization). |
| Animal Disease Models | For pre-clinical evaluation of biomaterial safety and efficacy in a physiologically relevant environment. | Critical-sized calvarial defect in rats (bone regeneration); full-thickness skin wound in mice (wound healing) [77]. |
The comparative analysis presented here underscores that no single biomaterial class is superior for all applications. The optimal choice is a deliberate decision based on a hierarchy of evidence, from fundamental material properties to robust clinical data. Key differentiators include the tunable versatility of polymers, the strength of metals, the bone-integration capacity of ceramics, and the tissue-like biocompatibility of hydrogels. The future of biomaterials lies in the intelligent design of composite and hybrid systems that transcend the limitations of individual classes, and in the adoption of data-driven approaches like machine learning to efficiently navigate the vast design space. As the field progresses, a steadfast commitment to evidence-based research will be paramount in translating innovative biomaterial solutions from the laboratory into safe and effective clinical products.
GRADE and other Systems for Assessing the Strength of Recommendations
In biomaterial research and drug development, the translation of preclinical and clinical evidence into reliable recommendations is paramount. Frameworks for assessing the quality of evidence and the strength of recommendations provide critical structure for this process, enhancing transparency, consistency, and rigor in evaluating scientific data [96] [97]. The Grading of Recommendations Assessment, Development, and Evaluation (GRADE) system has emerged as a internationally recognized methodology for this purpose, though several alternative approaches exist [96]. This guide objectively compares GRADE with other systems, focusing on their application in biomaterial research studies where evidence may originate from diverse study designs including randomized trials, observational studies, and in vitro experiments.
GRADE's primary function is to systematically assess the certainty in evidence (also known as quality of evidence or confidence in effect estimates) and to determine the strength of recommendations in healthcare and related research fields [96]. The system emphasizes the importance of considering not only the strengths and weaknesses of the evidence itself, but also values, preferences, and practical considerations when making recommendations [97]. For biomaterial researchers, this structured approach offers a mechanism to evaluate the collective evidence supporting a material's safety, efficacy, and performance.
The GRADE Approach: Framework and Components
Core Structure of GRADE
The GRADE approach operates through a structured process that separates the assessment of evidence quality from the formulation of recommendations. It provides a comprehensive framework for acknowledging the importance of outcomes of alternative management strategies, incorporating patient and public values and preferences, and applying explicit criteria for both downgrading and upgrading certainty in evidence [96]. This transparent process is particularly valuable in biomaterial research, where multidisciplinary teams must integrate diverse forms of evidence.
GRADE classifies the quality of evidence into four distinct levels [96]:
- High certainty: The research provides a lot of confidence that the true effect lies close to the estimated effect.
- Moderate certainty: The research provides moderate confidence in the estimated effect, but the true effect could be substantially different.
- Low certainty: The research provides limited confidence in the estimated effect, and the true effect might be substantially different from the estimated effect.
- Very low certainty: The research provides very little confidence in the estimated effect, and the true effect is likely substantially different from the estimated effect.
Determining Strength of Recommendations
In the GRADE framework, recommendations are categorized as either strong or weak (conditional) based on a transparent evaluation of multiple factors [96]. The direction and strength of a recommendation depend on the balance between desirable and undesirable consequences of following a course of action [96]. The certainty associated with this trade-off fundamentally determines the recommendation strength.
The GRADE system utilizes several criteria to determine the strength of recommendations [96]:
- Problem importance: The burden of disease, prevalence, or baseline risk of the healthcare issue being addressed.
- Quality of evidence: The overall confidence in the estimated effects across critical outcomes.
- Values and preferences: The importance, variability, and uncertainty associated with how affected individuals value the outcomes.
- Benefits and harms: The evaluation of absolute effects and importance of both beneficial and harmful outcomes.
- Resource implications: The cost-effectiveness and resource intensity of an intervention.
- Equity, acceptability, and feasibility: Considerations of how an option affects equity, its acceptability to stakeholders, and practical implementation feasibility.
Alternative Assessment Systems and Comparative Analysis
Alternative Systems and Methodologies
While GRADE is widely adopted, other methodologies exist for assessing evidence and recommendations. Understanding these alternatives provides context for selecting the most appropriate framework for specific research needs.
The Evidence-based Practice Center (EPC) Summary of Findings (SoF) table presents systematic review results in a concise, explicit format. Research has compared alternative SoF table formats against GRADE SoF tables to evaluate understanding, accessibility, satisfaction, and preference among systematic review users [98]. These comparisons are particularly relevant for biomaterial researchers communicating findings to diverse audiences including clinicians, regulators, and industry partners.
Trial Sequential Analysis (TSA) has been advocated as a supplementary methodology for a more reliable assessment of imprecision, which is one of the key domains in evidence evaluation [99]. TSA is used to control for random errors (imprecision) in meta-analyses, especially those with sparse data, by calculating a required information size and adjusting statistical thresholds to account for repetitive testing of accumulating data [99]. This method provides a more formal evaluation of imprecision than the qualitative rules of thumb sometimes employed in GRADE assessments.
Experimental Comparisons: GRADE vs. Alternative Formats
A three-arm, randomized, controlled, non-inferiority trial directly compared the standard GRADE Summary of Findings table with two alternative SoF table formats [98]. The primary objective was to investigate whether an alternative GRADE SoF table or an EPC SoF table was non-inferior to the current GRADE SoF table in terms of users' understanding of the information presented, particularly for descriptive findings [98].
The study recruited researchers, clinical practice guideline developers, policy-makers, and knowledge transfer professionals. Data was collected electronically at baseline and after randomization. The trial established a non-inferiority margin of 10%, meaning non-inferiority would be declared if the difference in the proportion of participants who understood the information displayed in the alternative SoF tables was 10% or less compared to the standard GRADE format [98].
Another cross-sectional study of 100 Cochrane reviews compared the assessment of imprecision using GRADE versus TSA [99]. The study found that only 13% of reviews clearly stated the criteria they applied to assess imprecision, with the most common dimensions being the 95% width of confidence intervals and optimal information size [99]. When the study authors re-analyzed imprecision following the GRADE Handbook guidelines, they found that 64% of outcomes warranted downgrading due to imprecision, compared to the 48% originally downgraded by review authors [99]. TSA was even more conservative, downgrading 69% of outcomes due to imprecision [99].
Table 1: Comparison of Agreement in Imprecision Assessment Between Methods
| Comparison Method | Agreement Level (Kappa Statistic) | 95% Confidence Interval |
|---|---|---|
| Review authors' GRADE vs. Handbook GRADE | 0.43 (Moderate) | 0.23 to 0.58 |
| Review authors' GRADE vs. TSA | 0.43 (Moderate) | 0.21 to 0.57 |
| Handbook GRADE vs. TSA | 0.66 (Substantial) | 0.49 to 0.79 |
Systematic Comparison of Framework Features
Table 2: Comparative Analysis of Evidence Assessment System Features
| Feature | GRADE | Trial Sequential Analysis (TSA) | Evidence-based Practice Center (EPC) SoF |
|---|---|---|---|
| Primary Purpose | Assess evidence certainty and strength of recommendations [96] [97] | Control random errors in meta-analysis; assess imprecision [99] | Present systematic review results concisely [98] |
| Evidence Certainty Levels | High, Moderate, Low, Very Low [96] | Not a primary feature (focuses on imprecision) | Varies by implementation |
| Recommendation Strength | Strong or Weak (Conditional) [96] | Not applicable | Not applicable |
| Key Assessment Domains | Risk of bias, imprecision, inconsistency, indirectness, publication bias [97] [99] | Imprecision (random error) | Dependent on the specific systematic review methodology |
| Handling of Imprecision | Based on optimal information size, confidence intervals, and effect thresholds [99] | Diversity-adjusted required information size (DARIS), sequential monitoring boundaries [99] | Varies by implementation |
| Strengths | Comprehensive, transparent, widely endorsed [96] | Provides more rigorous, quantitative assessment of imprecision [99] | Format designed for clarity and user understanding [98] |
| Limitations | Subjective application of imprecision criteria; variability in use [99] | Focuses primarily on imprecision, not other domains | Limited to presentation format rather than comprehensive assessment |
Experimental Protocols and Assessment Methodologies
Protocol for Assessing Imprecision Using GRADE
The GRADE methodology provides specific guidance for assessing imprecision, a key dimension in evaluating confidence in evidence [99]. The following protocol outlines the steps for proper evaluation of imprecision:
- Define the Outcome and Effect Measure: Clearly specify the dichotomous or continuous outcome and the appropriate effect measure (e.g., risk ratio, odds ratio, mean difference).
- Calculate Optimal Information Size (OIS): Determine the OIS using an alpha of 0.05, beta of 0.20 (for 80% power), and an a priori anticipated intervention effect. The intervention effect should be based on a clinically important effect threshold, which can be defined by the researchers or using a default threshold such as a 25% relative risk reduction if no specific threshold is available [99].
- Apply Downgrading Criteria:
- If the OIS criterion is not met, rate down for imprecision, unless the sample size is very large (at least 2000 patients, and perhaps 4000) [99].
- If the OIS criterion is met and the 95% confidence interval excludes no effect (e.g., risk ratio of 1.0), do not rate down for imprecision.
- If the OIS criterion is met, and the 95% confidence interval overlaps no effect, rate down for imprecision if the confidence interval fails to exclude important benefit or important harm [99].
Protocol for Trial Sequential Analysis
Trial Sequential Analysis provides a complementary methodology for a more rigorous assessment of imprecision [99]. The protocol involves:
- Data Extraction and Preparation: Extract data from all trials included in the meta-analysis for the specific outcome of interest.
- Cumulative Meta-Analysis: Sequentially add each trial to the analysis by publication year, creating a series of cumulative time points.
- Calculate Diversity-Adjusted Required Information Size (DARIS): Determine the DARIS using an alpha of 0.05, beta of 0.20, and a relative risk reduction as defined by the researchers or a default threshold (e.g., 25%). When using a random-effects model, adjust the required information size to account for between-trial variability (diversity, D²) [99].
- Construct Trial Sequential Monitoring Boundaries: Apply the Lan-DeMets trial sequential monitoring boundaries based on O'Brien-Fleming α-spending function [99].
- Plot Cumulative Z-Curve and Interpret Results: Calculate and plot the cumulative Z-curve (the series of Z-statistics after each consecutive trial) against the monitoring boundaries. Interpretation depends on whether the Z-curve crosses the trial sequential monitoring boundaries for benefit, harm, or futility [99].
Experimental Workflow for Framework Comparison
The following diagram illustrates the experimental workflow used in comparative studies of evidence assessment frameworks:
Table 3: Essential Reagents and Tools for Evidence Assessment
| Tool/Resource | Function/Purpose | Application Context |
|---|---|---|
| GRADE Handbook | Provides comprehensive guidance on applying GRADE methodology, including detailed criteria for rating evidence certainty and strength of recommendations [99]. | Essential reference for systematic review teams and guideline developers implementing GRADE. |
| GRADEpro Software | Software application that facilitates use of the GRADE approach, allows development of summary tables, and contains the GRADE handbook [96]. | Practical tool for creating Summary of Findings tables and evidence profiles; free for non-profit organizations. |
| Trial Sequential Analysis (TSA) Software | Software (v 0.9.5.5 Beta) used to perform TSA to control for random errors in meta-analyses [99]. | Used as a supplement to GRADE for more rigorous assessment of imprecision in meta-analyses. |
| Cochrane Systematic Reviews | High-quality systematic reviews that often employ GRADE methodology for evidence assessment [99]. | Source of evidence for guideline development and exemplars of methodology application. |
| Optimal Information Size (OIS) Calculator | Tool for calculating the required sample size in meta-analyses to detect or reject a specific intervention effect [99]. | Critical for proper application of GRADE's imprecision domain and for planning TSA. |
Application in Biomaterial Research and Limitations
Critical Considerations for Biomaterial Research
When applying evidence assessment frameworks like GRADE in biomaterial research, several field-specific considerations emerge. The standard GRADE approach has faced criticism when applied to certain research domains, including nutritional science, dietary, lifestyle, and environmental exposure research [96]. This criticism stems from GRADE's initial positioning that only allows randomized controlled trials to be rated as high-quality evidence, while automatically rating observational studies as low-quality due to their potential for confounding [96].
This limitation is particularly relevant to biomaterial research, where:
- Long-term effects of biomaterials and medical devices often cannot be practically or ethically studied using RCTs.
- Slowly progressing outcomes (such as biodegradation or chronic inflammatory responses) may be more appropriately studied through observational designs.
- Early-stage biomaterial development relies heavily on preclinical (in vitro and animal) studies, which present challenges for traditional evidence grading systems.
The criticism suggests that automatically dismissing observational evidence fails to acknowledge the strengths of well-designed observational studies and does not reflect the key limitations that RCTs themselves have for evaluating long-term effects [96]. For example, studying a slowly progressing process like atherosclerosis through RCTs presents significant challenges that observational studies may better address [96].
Integration of Diverse Evidence Types
Biomaterial research often involves integrating diverse types of evidence, from in vitro studies to animal models to human trials. The GRADE framework primarily focuses on evidence from human studies, creating a need for complementary approaches to assess preclinical evidence. Biomaterial researchers should consider:
- Developing domain-specific modifications to standard evidence assessment frameworks
- Creating transparent systems for integrating mechanistic evidence from laboratory studies with clinical outcomes
- Establishing biomaterial-specific criteria for what constitutes a "large effect" that might upgrade evidence from observational studies
The GRADE framework provides a comprehensive, transparent system for assessing evidence certainty and strength of recommendations, with wide adoption across healthcare organizations worldwide [96]. Experimental comparisons show that while GRADE remains the dominant approach, supplementary methodologies like Trial Sequential Analysis can provide more rigorous assessment of specific domains like imprecision [99]. Alternative presentation formats like modified Summary of Findings tables may also improve understanding and accessibility for end users [98].
For biomaterial researchers, effectively utilizing these frameworks requires careful consideration of field-specific challenges, particularly the heavy reliance on observational studies and preclinical evidence. Future developments in evidence assessment will likely include:
- Enhanced methodologies for integrating diverse evidence types
- Domain-specific modifications for specialized research areas
- Improved tools for visualizing and communicating evidence assessments
- Greater integration of artificial intelligence to assist in the evidence assessment process
By understanding the comparative strengths and limitations of different assessment frameworks, biomaterial researchers can more effectively evaluate their own research and critically appraise the growing body of evidence in this rapidly advancing field.
The translation of biomaterials from laboratory research to clinical applications requires a rigorous, evidence-based approach to evaluate their performance. Evidence-based biomaterials research (EBBR) utilizes systematic reviews and meta-analyses to translate vast amounts of research data into validated scientific evidence, establishing clear levels of evidence for biomaterial properties and performance [4]. This framework is particularly crucial for biodegradable metals in orthopedic applications, where materials must satisfy competing requirements: sufficient mechanical strength to bear physiological loads, controlled degradation rates to match bone healing timelines, and excellent biocompatibility to promote osteogenesis without causing adverse reactions [100]. Magnesium (Mg) alloys have emerged as promising candidates for bone fixation devices due to their biocompatibility, elastic modulus closer to natural bone than other metals, and ability to degrade in the biological environment while releasing osteopromotive ions [100]. However, the key challenge lies in simultaneously achieving a yield strength >200 MPa, elongation > 10%, and a controlled degradation rate <0.5 mm/year—benchmarks that pure magnesium fails to meet [100]. This comparison guide employs an evidence-based framework to objectively evaluate the performance of advanced Mg-Sr-Mn alloys against these critical clinical benchmarks and other biodegradable metal systems.
Performance Benchmarking of Biomaterials
Comparative Analysis of Biodegradable Metals
Table 1: Performance Comparison of Biodegradable Metals for Orthopedic Applications
| Material System | Yield Strength (MPa) | Ultimate Tensile Strength (MPa) | Elongation (%) | Degradation Rate (mm/year) | Elastic Modulus (GPa) | Key Advantages | Principal Limitations |
|---|---|---|---|---|---|---|---|
| Mg-0.3Sr-0.4Mn (SM04) [100] | 205 | 242 | Data Not Provided | 0.39 | 41-45 | Optimal strength-degradation balance, high ALP activity (2.46x increase) | Performance sensitive to precise composition |
| Pure Mg [100] | Insufficient for load-bearing sites | Insufficient for load-bearing sites | Data Not Provided | Too rapid | 41-45 | Excellent biocompatibility, promotes bone regeneration | Rapid degradation leads to premature mechanical failure |
| Iron (Fe) [100] | 200-250 | Data Not Provided | Data Not Provided | Excessively slow (~0.01) | ~211.4 | High mechanical strength | Stress shielding, incomplete absorption after healing |
| Zinc (Zn) [100] | 80-120 | Data Not Provided | Data Not Provided | Moderate | ~90 | Moderate degradation rate | Low strength inadequate for load-bearing sites |
| Mg-0.3Sr (SM0) [100] | ~160 | ~217 | Data Not Provided | 0.85 | 41-45 | Base for comparison, Sr addition refines grains | Lower strength and higher degradation vs. SM04 |
| Rolled Mg-2Sr [100] | ~150 | 213 | ~3 | 0.37 | 41-45 | Good ultimate tensile strength | Yield strength and elongation below clinical benchmarks |
In-Depth Analysis of Mg-Sr-Mn Alloy Series
Table 2: Detailed Properties of Extruded Mg-Sr-Mn Alloys with Varying Mn Content
| Alloy Designation | Mn Content (wt.%) | Average Grain Size (µm) | Yield Strength (MPa) | Ultimate Tensile Strength (MPa) | Corrosion Rate (mm/year) | Cell Viability | Osteogenic Performance |
|---|---|---|---|---|---|---|---|
| SM0 [100] | 0 | 7.43 | ~160 | ~217 | 0.85 | >90% | Baseline (1x ALP activity) |
| SM04 [100] | 0.4 | 4.42 | 205 | 242 | 0.39 | >90% | 2.46x higher ALP activity |
| SM12 [100] | 1.2 | 3.03 | Data Not Provided | Data Not Provided | Data Not Provided | Data Not Provided | Data Not Provided |
| SM20 [100] | 2.0 | 2.77 | Data Not Provided | Data Not Provided | Data Not Provided | Data Not Provided | Data Not Provided |
The data reveals that the Mg-0.3Sr-0.4Mn (SM04) alloy demonstrates optimal comprehensive performance, achieving a 28% increase in yield strength and an 11.5% increase in ultimate tensile strength compared to the binary Mg-0.3Sr (SM0) alloy, while simultaneously reducing the corrosion rate by 54% [100]. This aligns with the clinical benchmark for yield strength (>200 MPa) and degradation rate (<0.5 mm/year). Notably, excessive Mn addition beyond 0.4 wt.% (SM12, SM20) leads to continued grain refinement but may compromise corrosion resistance due to weakened basal texture, highlighting the need for precise compositional control [100].
Experimental Protocols for Biomaterial Evaluation
Material Fabrication and Microstructural Characterization
The Mg-Sr-Mn alloys were prepared with a fixed Sr content of 0.3 wt.% and varying Mn contents (0, 0.4, 1.2, and 2.0 wt.%) [100]. The alloys underwent extrusion processing to refine their microstructure and enhance mechanical properties. Microstructural characterization was performed using scanning electron microscopy (SEM) with energy-dispersive X-ray spectroscopy (EDS) mapping and transmission electron microscopy (TEM) to analyze phase distribution, precipitate formation, and elemental composition [100]. X-ray diffraction (XRD) was utilized to identify present phases, confirming α-Mg and Mg17Sr2 phases in all alloys, with α-Mn peaks becoming detectable at higher Mn concentrations [100]. Electron backscatter diffraction (EBSD) was employed to generate inverse pole figures and analyze grain size, orientation, and the distribution of low-angle grain boundaries, which indicated that dynamic recrystallization occurred predominantly through a continuous dynamic recrystallization mechanism [100].
Mechanical and Corrosion Testing Protocols
Tensile testing was conducted to determine yield strength, ultimate tensile strength, and elongation according to standardized methods. In vitro biodegradation behavior was evaluated by immersing samples in simulated physiological fluids, with corrosion rates calculated based on hydrogen evolution or mass loss measurements [100]. The study specifically noted that a key challenge is achieving a yield strength >200 MPa, elongation >10%, and a degradation rate <0.5 mm/year, benchmarks against which the alloys were evaluated [100].
Biocompatibility and Osteogenic Potential Assessment
Biocompatibility was assessed using cell viability assays (e.g., MTT assay) with osteoblast cell lines, where viability exceeding 90% was maintained for the optimal SM04 alloy [100]. Osteogenic differentiation was quantified by measuring alkaline phosphatase (ALP) activity, a key early marker of osteoblast function. The SM04 alloy demonstrated a 2.46-fold higher ALP activity compared to the SM0 alloy, indicating significantly enhanced osteogenic potential [100].
Performance Relationships in Mg-Sr-Mn Alloys
Property Interplay in Mg Alloy Design
The Scientist's Toolkit: Essential Research Reagents and Equipment
Table 3: Essential Research Tools for Biomaterials Development and Evaluation
| Tool/Reagent Category | Specific Examples | Primary Function in Research | Key Manufacturers/Providers |
|---|---|---|---|
| Mechanical Testing Systems | Mach-1 Multiaxial Tester | Performs compression, tension, bending, shear, friction, torsion and 3D indentation mapping of tissues and biomaterials [101]. | Biomomentum [101] |
| Biomaterial Fabrication | RESOMER bioresorbable polymers | Provides bioresorbable polymers for implant fabrication and comparative studies [101]. | Evonik [101] |
| Surface Analysis Instruments | Contact Angle Goniometers, Surface Tensiometers | Measures surface tension, contact angle, wettability, surface energy, and adhesion properties [101]. | DataPhysics Instruments [101] |
| Advanced Microscopy | Table-top Transmission Electron Microscopes (TEM) | Enables high-contrast imaging of microstructures and precipitate distribution in biomaterials [101]. | Delong Instruments [101] |
| Atomic Force Microscopy | Bio AFMs | Provides quantitative live-cell mechanical property mapping and high-resolution molecular imaging [101]. | Bruker [101] |
| Biometerials Testing & Culture | Bioprinters, Bioinks, Mechanical Testers | Enables 3D bioprinting of tissue constructs and specialized mechanical testing of biomaterials under physiological conditions [101]. | Allevi by 3D Systems, CellScale [101] |
| Calcium Phosphate Materials | Custom calcium phosphate powders, granules, cement powders | Serves as reference materials or components in comparative studies of bioactive bone graft substitutes [101]. | CaP Biomaterials [101] |
| Research Databases & Publications | Acta Biomaterialia, ACS Publications | Provides peer-reviewed research data, systematic reviews, and foundational knowledge for evidence-based research [101]. | Elsevier, ACS Publications [101] |
The field of biomaterials has undergone a revolutionary transformation, evolving from first-generation inert materials that simply provided structural support to advanced bioactive systems designed to actively direct biological responses and regeneration processes [102]. This progression necessitates a robust framework for evaluating and comparing the effectiveness of these material technologies. Evidence-based biomaterials research (EBBR) provides this essential methodology, using systematic approaches to translate vast research data into validated scientific evidence [18]. Within this hierarchy of evidence, in vitro studies provide fundamental mechanistic insights, animal models offer preclinical performance data, and human clinical trials deliver the most clinically relevant effectiveness measures.
This guide employs this evidence-based framework to objectively compare traditional and advanced biomaterials across critical application areas. We present structured experimental data and detailed methodologies to facilitate informed material selection for researchers and development professionals working toward therapeutic translation.
Comparative Analysis in Bone Regeneration
Performance Data and Experimental Evidence
The quest for effective bone regeneration strategies highlights the distinct performance characteristics of traditional metals, bioactive ceramics, and emerging nanocomposites.
Table 1: Comparative Performance of Biomaterials for Bone Regeneration
| Material Type | Key Formulations | Osseointegration Capability | Mechanical Properties | Degradation Profile | Evidence Level |
|---|---|---|---|---|---|
| Traditional Metals | Titanium (Ti), Ti6Al4V, 316L Stainless Steel [102] [103] | Moderate; often requires surface modification to enhance bonding [102] | High strength & durability (Excellent for load-bearing) [103] | Non-degradable / Overstable (may require removal) [102] | Clinical/Human (Established long-term use) |
| Bioactive Ceramics | Hydroxyapatite (HAp), Bioactive Glasses [104] [103] | High; osteoconductive, bonds directly to bone [103] | Brittle, low tensile strength (Good compression) [103] | Slow to non-degradable [103] | Pre-Clinical / Clinical |
| Advanced Nanocomposites | BC/MXene/HAp/MNPs, HAp-Polymer Scaffolds [102] [104] | Very High; designed to be osteoinductive [104] | Tunable to match bone mechanics [104] | Controllable degradation to match tissue growth [102] | In Vitro / Pre-Clinical |
Experimental Protocols for Bone Regeneration Studies
Protocol for Evaluating Osseointegration of Titanium Implants:
- Implant Preparation: Titanium surfaces are roughened via grit-blasting and acid-etching or coated with bioactive molecules (e.g., fibrinogen using polydopamine chemistry) to enhance cellular response [102].
- Animal Model: Implants are placed in critical-sized bone defects (e.g., canine mandibular defect or rat calvarial defect) [102] [105].
- Analysis Timeline: Animals are sacrificed at 4, 8, and 12 weeks for histological analysis.
- Outcome Measures: Histomorphometry to quantify bone-to-implant contact (BIC), push-out tests for mechanical integration strength, and micro-CT scanning to assess bone volume and trabecular architecture around the implant [102].
Protocol for Testing Bioactive Nanocomposites:
- Scaffold Fabrication: A composite scaffold of Bacterial Cellulose (BC), Hydroxyapatite (HAp), and Metallic Nanoparticles (MNPs) is produced using 3D printing or freeze-drying to create a porous structure [104].
- In Vitro Assessment: Scaffolds are seeded with human Mesenchymal Stem Cells (hMSCs). Osteogenic differentiation is quantified by measuring alkaline phosphatase (ALP) activity at 7 and 14 days, and mineral deposition (via Alizarin Red staining) at 21 days [104] [106].
- In Vivo Assessment: Scaffolds are implanted in segmental bone defect models. The primary outcome is the amount of new bone formation within the scaffold pores, assessed longitudinally using micro-CT and confirmed with post-mortem histology [104].
Signaling Pathways in Bone Regeneration
Advanced biomaterials are designed to actively modulate specific signaling pathways to enhance bone healing, a key differentiator from traditional inert materials.
Comparative Analysis in Skin and Soft Tissue Wound Healing
Performance Data and Experimental Evidence
The primary challenge in wound healing is achieving regenerative tissue repair without scar formation, a key area where advanced biomaterials show significant promise.
Table 2: Comparative Performance of Biomaterials for Wound Healing
| Material Type | Key Formulations | Scar Formation | Re-epithelialization Rate | Immunomodulatory Capacity | Evidence Level |
|---|---|---|---|---|---|
| Traditional Dressings | Gauze, Silicone Sheets, Polyurethane Films [107] [103] | High (Promotes scar tissue) [107] | Slow | Passive barrier; minimal immune interaction [108] | Clinical/Human |
| Natural Polymer Hydrogels | Chitosan, Alginate, Collagen-based [103] [108] | Moderate | Moderate | Innate anti-inflammatory properties (e.g., Chitosan) [108] | Pre-Clinical / Clinical |
| Advanced Immunomodulatory Scaffolds | Electrospun nanofibers with anti-inflammatory agents (e.g., CP copolymers) [103] [108] | Low (Promotes regeneration) [107] [108] | Rapid | Active macrophage polarization (M1 to M2 phenotype) [108] | In Vitro / Pre-Clinical |
Experimental Protocols for Wound Healing Studies
Protocol for Testing Immunomodulatory Biomaterials:
- Biomaterial Fabrication: A chitosan-poloxamer 123 (CP) copolymer hydrogel is synthesized and loaded with anti-inflammatory cytokines (e.g., IL-4 or IL-10) [103] [108].
- In Vitro Macrophage Culture: Human monocytes (THP-1 cells) are differentiated into M0 macrophages and then exposed to the biomaterial leachates. Macrophage polarization is analyzed via flow cytometry for M1 (CD80) and M2 (CD206) surface markers, and by quantifying pro-inflammatory (TNF-α) vs. anti-inflammatory (IL-10) cytokine secretion [108].
- In Vivo Wound Model: Full-thickness excisional wounds are created on diabetic (db/db) mice. The hydrogel is applied, and wounds are documented daily. Tissue is harvested at days 7, 14, and 21 for histology (H&E staining to measure epithelial gap, Masson's Trichrome for collagen organization) and immunofluorescence to identify M1/M2 macrophages in the wound bed [108].
The Scientist's Toolkit: Key Reagents and Materials
Table 3: Essential Research Reagents for Biomaterials Evaluation
| Reagent/Material | Function in Experimental Protocols | Example Application |
|---|---|---|
| Mesenchymal Stem Cells (hMSCs) | Multipotent progenitor cells used to assess osteogenic and chondrogenic differentiation potential of biomaterials. | Seeded on bone scaffolds to test new tissue formation [107] [106]. |
| THP-1 Cell Line | A human monocytic cell line that can be differentiated into macrophages for standardized in vitro immunomodulation tests. | Used to screen biomaterials for their ability to polarize macrophages to pro-healing M2 phenotype [108]. |
| Alkaline Phosphatase (ALP) Assay Kit | Quantifies ALP enzyme activity, an early marker of osteoblastic differentiation. | Measuring osteoinductivity of bone biomaterials after 7-14 days of cell culture [106]. |
| Alizarin Red S | A dye that binds to calcium deposits, used to visualize and quantify mineralized matrix formation in vitro. | Staining mineral nodules after 21-28 days of osteogenic culture to confirm bone nodule formation [106]. |
| ELISA Kits for Cytokines | Enzyme-linked immunosorbent assay kits for quantifying specific proteins (e.g., TNF-α, IL-6, IL-10) in cell culture supernatant. | Assessing the inflammatory response (pro-inflammatory vs. anti-inflammatory) elicited by a biomaterial [108]. |
| Polydopamine Coating | A versatile mussel-inspired surface chemistry for priming material surfaces for further functionalization. | Improving the bonding of proteins (e.g., fibrinogen) to metal surfaces like Ti6Al4V to enhance biological sealing [102]. |
Methodological Considerations and Evidence Grading
Workflow for Evidence-Based Biomaterial Assessment
A systematic approach is critical for translating basic research findings into clinically applicable evidence.
Optimizing Research with Design of Experiments and Machine Learning
The complexity of advanced biomaterials, which often involve multiple interacting components (e.g., polymers, ceramics, ions), requires sophisticated experimental design beyond traditional one-factor-at-a-time approaches. Design of Experiments (DoE) is a statistical methodology that systematically varies all relevant input factors simultaneously to model their relationships with output responses (e.g., scaffold stiffness, degradation rate) [109]. This allows for the identification of optimal material compositions with a reduced number of experimental runs, saving time and resources.
Emerging methodologies now combine DoE with Machine Learning (ML). ML algorithms, such as artificial neural networks, can process complex, high-dimensional data (including images from histology or micro-CT) to predict biomaterial performance and uncover non-intuitive relationships between material composition and biological outcomes that might be missed by conventional analysis [109]. This powerful combination is accelerating the rational design and optimisation of next-generation biomaterials.
The comparative analysis presented herein demonstrates a clear efficacy gradient: advanced biomaterials consistently outperform traditional alternatives in key biological functionalities such as osseointegration, immunomodulation, and promotion of regenerative healing. The transition from inert biocompatibility to bioactive design represents a paradigm shift in biomaterials science.
Future progress hinges on several key frontiers. First, the integration of machine learning and data-driven design will move the field from empirical discovery to predictive material development [17] [109]. Second, a deeper understanding of personalized immunomodulation is needed to tailor biomaterials to a patient's specific immune profile, thereby improving outcomes and reducing rejection [108]. Finally, overcoming translational challenges requires a full-chain approach that aligns fundamental research with regulatory pathways and manufacturing scalability from the outset [18] [102]. By adhering to evidence-based principles and leveraging these emerging technologies, researchers can continue to develop increasingly sophisticated biomaterial solutions that address unmet clinical needs.
Conclusion
The adoption of a structured, evidence-based approach is paramount for advancing the field of biomaterials. By understanding and applying the hierarchy of evidence, researchers can more effectively translate basic research into safe and effective clinical products. Methodologies like systematic reviews and meta-analyses provide a powerful means to synthesize existing data, while emerging tools like AI offer new avenues for optimization and prediction. Future progress hinges on rigorous validation, interdisciplinary collaboration, and a continued focus on generating high-quality evidence that satisfies both scientific and regulatory requirements. This will ultimately accelerate the development of next-generation biomaterials, enhancing predictive and personalized healthcare outcomes.
References
- 1. Systematic Comparison of Biomaterialsâ€Based Strategies ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11468758/]
- 2. A Comparison of Natural and Synthetic Biomaterials in ... [https://research-archive.org/index.php/rars/preprint/view/3020]
- 3. Biomaterials by design: Harnessing data for future ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8628044/]
- 4. Evidence-based biomaterials research [https://www.sciencedirect.com/science/article/pii/S2452199X22001840]
- 5. Evidence-based biomaterials research [https://pubmed.ncbi.nlm.nih.gov/35633843/]
- 6. A Blueprint for Translational Regenerative Medicine - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7610850/]
- 7. Levels of evidence in research [https://scientific-publishing.webshop.elsevier.com/research-process/levels-of-evidence-in-research/]
- 8. Levels of Evidence - Evidence-Based Medicine [https://guides.library.stonybrook.edu/evidence-based-medicine/levels_of_evidence]
- 9. Evidence-Based Practice: Levels of Evidence [https://libguides.mskcc.org/ebp/evidence]
- 10. Comparison of Printable Biomaterials for Use in Neural ... [https://www.mdpi.com/2073-4360/16/10/1426]
- 11. New Translational Roadmap Promises Faster, Safer AI in ... [https://engineering.purdue.edu/BME/AboutUs/News/2025/new-translational-roadmap-promises-faster-safer-ai-in-healthcare]
- 12. Understanding the Levels of Evidence in Medical Research [https://pmc.ncbi.nlm.nih.gov/articles/PMC12064251/]
- 13. A comprehensive guide to conduct a systematic review and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12366998/]
- 14. A Histological and Clinical Evaluation of Long-Term ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12470767/]
- 15. Whitlockite as a next-generation biomaterial for bone ... [https://www.sciencedirect.com/science/article/pii/S2212426825001757]
- 16. Material synthesis and design optimization of biomaterials for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11954756/]
- 17. Biomaterials by design: Harnessing data for future ... [https://www.sciencedirect.com/science/article/pii/S2590006421000739]
- 18. Evidence-based biomaterials research - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9122837/]
- 19. Primary versus secondary source of data in observational ... [https://bmcmedresmethodol.biomedcentral.com/articles/10.1186/s12874-018-0561-3]
- 20. Biomaterials Market Size to Hit USD 523.75 Billion by 2034 [https://www.precedenceresearch.com/biomaterials-market]
- 21. The FDA Approval Process Simplified: A 2025 Guide ... [https://ovasolutions.com/the-fda-approval-process-simplified-a-2025-guide-for-medtech-innovators/]
- 22. FDA use of Real-World Evidence in Regulatory Decision ... [https://www.fda.gov/science-research/real-world-evidence/fda-use-real-world-evidence-regulatory-decision-making]
- 23. Rethinking FDA's Accelerated Approval Pathway [https://www.mcguirewoods.com/client-resources/alerts/2025/1/rethinking-fdas-accelerated-approval-pathway-new-draft-guidances-and-implications-for-drug-companies/]
- 24. Quantitative Research Designs, Hierarchy of Evidence and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12056466/]
- 25. Next-Generation Biomaterials: Design Strategies, Clinical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12529143/]
- 26. Biomaterial Market Size & Growth Outlook 2023-2035 [https://www.futuremarketinsights.com/reports/biomaterials-market]
- 27. Challenges in Biomaterials' Clinical Translation [https://www.biomatdb.eu/2023/08/09/challenges-in-biomaterials-clinical-translation-navigating-innovation-and-complexity/]
- 28. Systematic Review vs Meta Analysis: Key Differences (2025) [https://paperguide.ai/blog/systematic-review-vs-meta-analysis/]
- 29. Systematic review and meta-analysis in clinical trials [https://www.sciencedirect.com/science/article/pii/S1357303925000775]
- 30. Methodological Standards for Conducting High-Quality ... [https://www.mdpi.com/2079-7737/14/8/973]
- 31. Building a synthesis-ready research ecosystem: fostering ... [https://bmcmedresmethodol.biomedcentral.com/articles/10.1186/s12874-025-02524-2]
- 32. A comparative study of screening performance between ... [https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-025-03138-w]
- 33. A systematic review and meta-analysis of diagnostic ... [https://www.nature.com/articles/s41746-025-01543-z]
- 34. Exploring the Role of Artificial Intelligence in Evidence Synthesis [https://pmc.ncbi.nlm.nih.gov/articles/PMC12419556/]
- 35. Biological responses to biomaterials: a review - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12068782/]
- 36. Biomaterial research using MP-SPR and QCMD [https://bionavis.com/applications/biomaterials/]
- 37. A systematic review on bioactive glass as a treatment for limb ... [https://boneandjoint.org.uk/Article/10.1302/2633-1462.610.BJO-2025-0084.R2]
- 38. Efficacy of bioactive glass versus traditional bone grafts in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12553929/]
- 39. Bioactive Glasses: Frontiers and Challenges - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4663244/]
- 40. From benchtop to bedside: Precision-engineered bioactive ... [https://www.sciencedirect.com/science/article/pii/S2590049825001080]
- 41. Bioactive Glass Market Size, Share & Growth Forecast, 2034 [https://www.gminsights.com/industry-analysis/bioactive-glass-market]
- 42. Bioactive glasses as bone substitutes for animal critical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12551993/]
- 43. Real-World Evidence [https://www.fda.gov/science-research/science-and-research-special-topics/real-world-evidence]
- 44. Real-World Evidence: A Primer - PMC - PubMed Central - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC9815890/]
- 45. Harnessing digital health technologies and real-world ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12290384/]
- 46. Post-Marketing Surveillance in 2025: Evolving Strategies ... [https://www.vitrana.com/post-marketing-surveillance-in-2025-evolving-strategies-for-real-world-safety-monitoring/]
- 47. Real-world data to support post-market safety and ... [https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-024-02659-0]
- 48. “Real†Evidence Generating Real Pre- and Post-Market ... [https://globalforum.diaglobal.org/issue/june-2024/real-evidence-generating-real-pre-and-post-market-benefits/]
- 49. Systematic review: a scientometric analysis of the status ... [https://www.nature.com/articles/s40494-025-01636-8]
- 50. A Scientometric Analysis and Visualization Discovery of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9450939/]
- 51. Biomaterials in Medicine: Current Trends and Future ... [https://www.sciencepublishinggroup.com/article/10.11648/j.ijbmr.20251301.13]
- 52. Scientometric analysis of trends in research ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12055652/]
- 53. Scientometric Analysis of Research Work on Mental ... [https://www.mdpi.com/2076-3387/15/11/450]
- 54. Scientometric Analysis and Visualization of Research ... [https://link.springer.com/article/10.1007/s44230-025-00089-3]
- 55. The scientometric analysis and visualization of sustainable ... [https://www.sciencedirect.com/science/article/pii/S2405844023081938]
- 56. A visualized and scientometric analysis of research trends ... [https://www.frontiersin.org/journals/public-health/articles/10.3389/fpubh.2022.928720/full]
- 57. A SCIENTOMETRIC ANALYSIS OF GLOBAL RESEARCH ... [https://tpmap.org/submission/index.php/tpm/article/view/1827]
- 58. Challenges and Pitfalls of Research Designs Involving ... [https://pubmed.ncbi.nlm.nih.gov/38892430/]
- 59. Challenges With the Development of Biomaterials for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6554598/]
- 60. Challenges and Pitfalls of Research Designs Involving ... [https://www.mdpi.com/1422-0067/25/11/6242]
- 61. Common mistakes in biostatistics - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11333961/]
- 62. Common Bioinformatics Data Analysis Mistakes [https://bigomics.ch/blog/experimental-design-and-data-analysis-common-bioinformatics-mistakes/]
- 63. Considering best practices in color palettes for molecular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9377702/]
- 64. Ten simple rules to colorize biological data visualization [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1008259]
- 65. Bridging the Gap: Translating Preclinical Biomarkers to ... [https://blog.crownbio.com/bridging-the-gap-translating-preclinical-biomarkers-to-clinical-success]
- 66. The Role of Artificial Intelligence in Biomaterials Science [https://pmc.ncbi.nlm.nih.gov/articles/PMC12526772/]
- 67. Mapping Biomaterial Complexity by Machine Learning - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12394815/]
- 68. Applied machine learning as a driver for polymeric ... [https://www.nature.com/articles/s41467-023-40459-8]
- 69. AI vs Predictive Analytics: Key Differences & Use Cases ... [https://julius.ai/articles/ai-vs-predictive-analytics]
- 70. 3 Predictions For Predictive AI In 2025 [https://www.forbes.com/sites/ericsiegel/2025/01/06/3-predictions-for-predictive-ai-in-2025/]
- 71. AI Model Comparison 2025: 9 Top Contenders Ranked ... [https://www.chatbench.org/ai-model-comparison/]
- 72. Machine Learning in Biomaterials, Biomechanics ... [https://link.springer.com/article/10.1007/s11831-024-10100-y]
- 73. Engineering the Immune Response to Biomaterials - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12097135/]
- 74. Smart biomaterials: as active immune modulators to shape ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1669399/full]
- 75. Immune Response to Extracellular Matrix Bioscaffolds [https://www.mdpi.com/2673-8449/5/3/28]
- 76. Knowledge map in biomaterials and related immune ... [https://www.frontiersin.org/journals/materials/articles/10.3389/fmats.2025.1534127/full]
- 77. Research Progress on Biomaterials with Immunomodulatory ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12412535/]
- 78. Advances in engineering immune–tumor microenvironments ... [https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-025-02479-4]
- 79. Current landscape and challenges in adjuvant and antigen ... [https://www.sciencedirect.com/science/article/pii/S2590136225001299]
- 80. High Throughput Screening - an overview [https://www.sciencedirect.com/topics/chemistry/high-throughput-screening]
- 81. Quantitative high-throughput screening data analysis [https://pmc.ncbi.nlm.nih.gov/articles/PMC4375054/]
- 82. Materials Informatics 2025-2035: Markets, Strategies, Players [https://www.idtechex.com/en/research-report/materials-informatics-2025/1096]
- 83. Materials Informatics Market Size to Hit USD ... [https://www.precedenceresearch.com/material-informatics-market]
- 84. Material Informatics Market to Grow at 20.80% CAGR Driven [https://www.globenewswire.com/news-release/2025/09/23/3154585/0/en/Material-Informatics-Market-to-Grow-at-20-80-CAGR-Driven-by-Rising-Adoption-of-AI-and-Machine-Learning.html]
- 85. Good Laboratory Practice Everything You Need to Know [https://testlabsuk.com/blog/a-guide-to-good-laboratory-practice-glp-everything-you-need-to-know/]
- 86. Good Laboratory Practices vs. Good Manufacturing Practices [https://www.eurofinsus.com/food-testing/resources/good-laboratory-practices-vs-good-manufacturing-practices-whats-the-difference/]
- 87. ISO 10993-1:2025 Updates —Top 10 Biological Evaluation ... [https://namsa.com/resources/blog/iso-10993-12025top-10-biological-evaluation-essentials-in-the-update/]
- 88. ISO 10993-1:2025 Publication Is Imminent [https://veranex.com/blog/iso-10993-1-2025-implications-med-device-innovators-manufacturers]
- 89. The Key Concepts of Biocompatibility Testing [https://pacificbiolabs.com/the-key-concepts-of-biocompatibility-testing/]
- 90. Polymeric Materials in Biomedical Engineering: A Bibliometric ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12608308/]
- 91. Biomaterials Market Report 2025-2034 | Trends [https://www.thebusinessresearchcompany.com/report/biomaterials-global-market-report]
- 92. Next-Generation Hydrogel Design: Computational ... [https://www.mdpi.com/2073-4360/17/10/1373]
- 93. Tailored polymeric hydrogels for regenerative medicine ... [https://www.sciencedirect.com/science/article/pii/S0378517325006556]
- 94. Hydrogel-based piezoelectric materials and devices for ... [https://www.sciencedirect.com/science/article/abs/pii/S0142961225006878]
- 95. Data-driven de novo design of super-adhesive hydrogels [https://www.nature.com/articles/s41586-025-09269-4]
- 96. approach - Wikipedia GRADE [https://en.wikipedia.org/wiki/GRADE_approach]
- 97. Introduction to the GRADE tool for rating certainty in ... [https://www.sciencedirect.com/science/article/pii/S2213398423002713]
- 98. Two alternatives versus the standard Grading of Recommendations ... [https://pubmed.ncbi.nlm.nih.gov/29362242/]
- 99. imprecision in Cochrane Assessing reviews: a comparison... systematic [https://systematicreviewsjournal.biomedcentral.com/articles/10.1186/s13643-018-0770-1]
- 100. Controllable degradation behavior, mechanical properties ... [https://www.nature.com/articles/s41529-025-00658-8]
- 101. Resource Guide | Society for Biomaterials (SFB) [https://biomaterials.org/about-society/resource-guide]
- 102. Recent advances in regenerative biomaterials - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9745784/]
- 103. An Overview of Biomaterials: Advancements and ... [https://www.azom.com/article.aspx?ArticleID=108]
- 104. Advanced biomaterials in tissue engineering: A critical ... [https://www.sciencedirect.com/science/article/abs/pii/S0001868625002453]
- 105. Experimental Research and Computational Analysis of Eco [https://www.mdpi.com/1996-1944/17/17/4269]
- 106. Essential molecular biology methods in biomaterials ... [https://www.explorationpub.com/Journals/ebmx/Article/101344]
- 107. Advanced biomaterials for regenerative medicine and their ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11634172/]
- 108. Advanced biomaterials in immune modulation: The future ... [https://www.sciencedirect.com/science/article/abs/pii/S0378517325008099]
- 109. Design of Experiments in the Advancement of Biomaterial [https://encyclopedia.pub/entry/40331]